1F6L
 
 | | VARIABLE LIGHT CHAIN DIMER OF ANTI-FERRITIN ANTIBODY | | Descriptor: | ANTI-FERRITIN IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Nymalm, Y, Kravchuk, Z, Salminen, T, Chumanevich, A.A, Dubnovitsky, A.P, Kankare, J, Pentikainen, O, Lehtonen, J, Arosio, P, Martsev, S, Johnson, M.S. | | Deposit date: | 2000-06-22 | | Release date: | 2002-10-23 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antiferritin VL homodimer binds human spleen ferritin with high specificity
J.STRUCT.BIOL., 138, 2002
|
|
3RG6
 
 | | Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco | | Descriptor: | RbcX protein, Ribulose bisphosphate carboxylase large chain | | Authors: | Bracher, A, Starling-Windhof, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2011-04-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3RTK
 
 | | Crystal structure of Cpn60.2 from Mycobacterium tuberculosis at 2.8A | | Descriptor: | 60 kDa chaperonin 2, MAGNESIUM ION | | Authors: | Shahar, A, Melamed-Frank, M, Kashi, Y, Adir, N. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The dimeric structure of the Cpn60.2 chaperonin of Mycobacterium tuberculosis at 2.8 A reveals possible modes of function.
J.Mol.Biol., 412, 2011
|
|
1FCG
 
 | | ECTODOMAIN OF HUMAN FC GAMMA RECEPTOR, FCGRIIA | | Descriptor: | PROTEIN (FC RECEPTOR FC(GAMMA)RIIA) | | Authors: | Maxwell, K.F, Powell, M.S, Garrett, T.P, Hogarth, P.M. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa.
Nat.Struct.Biol., 6, 1999
|
|
7Y8P
 
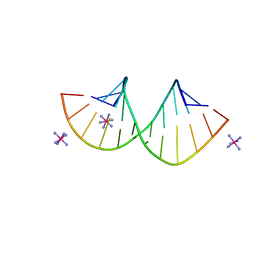 | | Crystal structure of 4'-selenoRNA duplex | | Descriptor: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | Authors: | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
3P8H
 
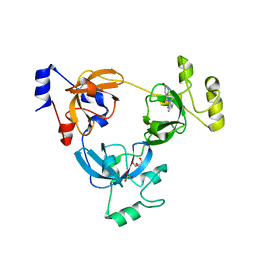 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
6HT5
 
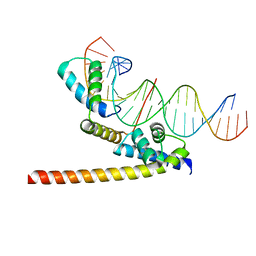 | | Oct4/Sox2:UTF1 structure | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*AP*GP*CP*AP*TP*AP*AP*CP*AP*AP*TP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*CP*AP*TP*TP*GP*TP*TP*AP*TP*GP*CP*TP*AP*GP*TP*GP*AP*AP*G)-3'), POU domain, ... | | Authors: | Vahokoski, J, Meusch, D, Groves, M, Pogenberg, V, Wilmanns, M. | | Deposit date: | 2018-10-03 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | Oct4/Sox2:UTF1 structure
To Be Published
|
|
3QTB
 
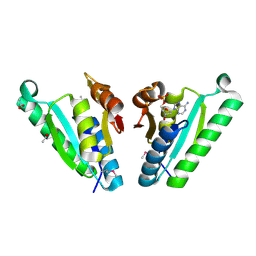 | | Structure of the universal stress protein from Archaeoglobus fulgidus in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, Uncharacterized protein | | Authors: | Tkaczuk, K.L, Shumilin, I.A, Chruszcz, M, Cymborowski, M, Xu, X, Di Leo, R, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
3RDQ
 
 | | Crystal structure of R7-2 streptavidin complexed with desthiobiotin | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
6K5P
 
 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ESS
 
 | | Structure-guided studies of the Holliday junction resolvase RuvX provide novel insights into ATP-stimulated cleavage of branched DNA and RNA substrates | | Descriptor: | Putative pre-16S rRNA nuclease | | Authors: | Thakur, M, Mohan, D, Singh, A.K, Agarwal, A, Gopal, B, Muniyappa, K. | | Deposit date: | 2021-05-11 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Novel insights into ATP-Stimulated Cleavage of branched DNA and RNA Substrates through Structure-Guided Studies of the Holliday Junction Resolvase RuvX.
J.Mol.Biol., 433, 2021
|
|
7EZL
 
 | | Rice L-galactose dehydrogenase (holo form) | | Descriptor: | L-galactose dehydrogenase, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Momma, M, Fujimoto, Z. | | Deposit date: | 2021-06-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of rice L-galactose dehydrogenase
To Be Published
|
|
7EZI
 
 | |
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
3NNQ
 
 | | Crystal Structure of the N-terminal domain of Moloney murine leukemia virus integrase, Northeast Structural Genomics Consortium Target OR3 | | Descriptor: | ACETATE ION, N-terminal domain of Moloney murine leukemia virus integrase, ZINC ION | | Authors: | Guan, R, Xiao, R, Acton, T, Jiang, M, Roth, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-24 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | X-ray crystal structure of the N-terminal region of Moloney murine leukemia virus integrase and its implications for viral DNA recognition.
Proteins, 85, 2017
|
|
3RE6
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDX
 
 | | Crystal structure of ligand-free R7-2 streptavidin | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDM
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin/PEG | | Descriptor: | BIOTIN, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDU
 
 | | Crystal structure of R7-2 streptavidin complexed with PEG | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
8QEA
 
 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 96 fs structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-08-31 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QK7
 
 | | E167K RF2 on E. coli 70S release complex with UAA | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
4Q4M
 
 | | tRNA-Guanine Transglycosylase (TGT) in Complex with 6-Amino-4-phenyl-1,2-dihydro-1,3,5-triazin-2-one | | Descriptor: | 6-amino-4-phenyl-1,3,5-triazin-2(1H)-one, DIMETHYL SULFOXIDE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-04-15 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | 5-Azacytosines as a Novel Scaffold to Inhibit Z. mobilis TGT with Expected Improved Bioavailability and Synthetic Accessibility
To be Published
|
|
6K2E
 
 | | Crystal structure of cas2 | | Descriptor: | CRISPR/cas2 protein | | Authors: | Mo, X, Bi, M, Yuan, A.Y. | | Deposit date: | 2019-05-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into endonuclease CRISPR-associated Cas2 protein
To be published
|
|
8QFK
 
 | |
