6JMU
 
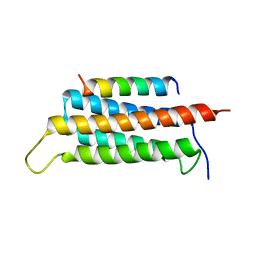 | | Crystal structure of GIT1/Paxillin complex | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
7VMB
 
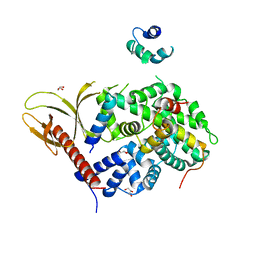 | |
4PI9
 
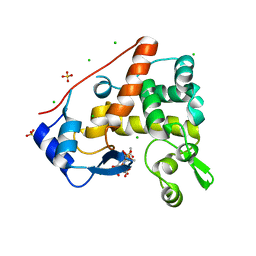 | | Crystal structure of S. Aureus Autolysin E in complex with muropeptide NAM-L-ALA-D-iGLU | | Descriptor: | (4R)-4-[[(2S)-2-[[(2R)-2-[(2R,3S,4R,5R,6R)-5-acetamido-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]oxypropanoyl]amino]propanoyl]amino]-5-azanyl-5-oxidanylidene-pentanoic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers.
IUCrJ, 4, 2017
|
|
4P6T
 
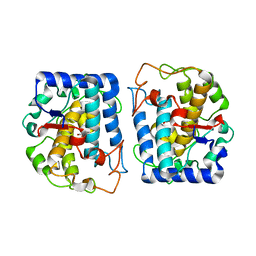 | | Crystal Structure of tyrosinase from Bacillus megaterium with p-tyrosol in the active site | | Descriptor: | 4-(2-hydroxyethyl)phenol, COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
8VSC
 
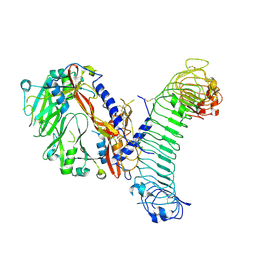 | | L-TGF-b1/GARP | | Descriptor: | Transforming growth factor beta activator LRRC32, Transforming growth factor beta-1 proprotein | | Authors: | Jin, M, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamic allostery drives autocrine and paracrine TGF-beta signaling.
Cell, 2024
|
|
2BQW
 
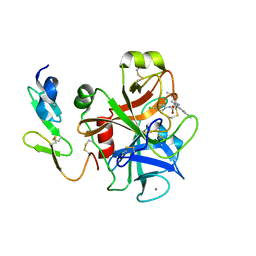 | | CRYSTAL STRUCTURE OF FACTOR XA IN COMPLEX WITH COMPOUND 45 | | Descriptor: | 1-{2-[(4-CHLOROPHENYL)AMINO]-2-OXOETHYL}-N-(1-ISOPROPYLPIPERIDIN-4-YL)-1H-INDOLE-2-CARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-28 | | Release date: | 2006-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
3LDH
 
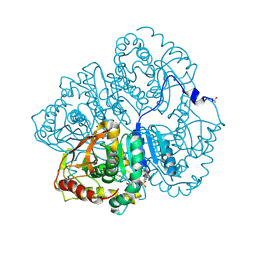 | | A comparison of the structures of apo dogfish m4 lactate dehydrogenase and its ternary complexes | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | White, J.L, Hackert, M.L, Buehner, M, Adams, M.J, Ford, G.C, Lentzjunior, P.J, Smiley, I.E, Steindel, S.J, Rossmann, M.G. | | Deposit date: | 1974-06-06 | | Release date: | 1977-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes.
J.Mol.Biol., 102, 1976
|
|
7OCV
 
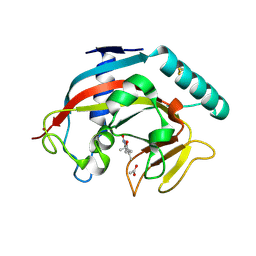 | | Human TNKS1 in complex with 3-[4-(1-Hydroxy-1-methyl-ethyl)-phenyl]-6-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one | | Descriptor: | 6-methyl-3-[4-(2-oxidanylpropan-2-yl)phenyl]-4~{H}-pyrrolo[1,2-a]pyrazin-1-one, ACETATE ION, Poly [ADP-ribose] polymerase, ... | | Authors: | Musil, D, Lehmann, M, Buchstaller, H.-P. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Optimization of a Screening Hit toward M2912, an Oral Tankyrase Inhibitor with Antitumor Activity in Colorectal Cancer Models.
J.Med.Chem., 64, 2021
|
|
7WEM
 
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | Descriptor: | ATP synthase subunit c | | Authors: | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
8VDL
 
 | | HB3VAR03 CIDRa1.4 domain with C7 Fab | | Descriptor: | C7 Heavy Chain, C7 Light Chain, HB3VAR03 CIDRa1.4 domain, ... | | Authors: | Hurlburt, N.K, Pancera, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Broadly inhibitory antibodies against severe malaria-associated PfEMP1 function through a structurally convergent mode of binding
To Be Published
|
|
8VA7
 
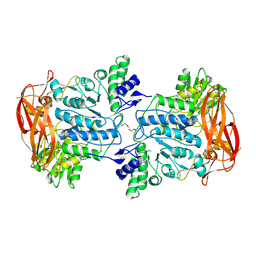 | | Crystal structure of CapGH3a enzyme retrieved from capybara gut metagenome | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycoside hydrolase family 3, ... | | Authors: | Martins, M.P, Vieira, P.S, Morais, M.A.B, Mandelli, F, Chinaglia, M, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
2BQ7
 
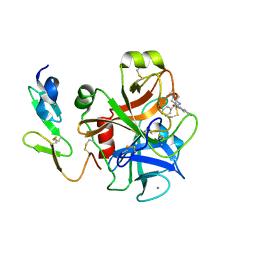 | | Crystal structure of factor Xa in complex with 43 | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, FACTOR XA, ... | | Authors: | Nazare, M, Will, D.W, Matter, H, Schreuder, H, Ritter, K, Urmann, M, Essrich, M, Bauer, A, Wagner, M, Czech, J, Laux, V, Wehner, V. | | Deposit date: | 2005-04-27 | | Release date: | 2006-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the Subpockets of Factor Xa Reveals Two Binding Modes for Inhibitors Based on a 2-Carboxyindole Scaffold: A Study Combining Structure-Activity Relationship and X-Ray Crystallography.
J.Med.Chem., 48, 2005
|
|
7OD0
 
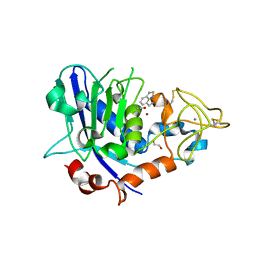 | | Mirolysin in complex with compound 9 | | Descriptor: | 1,2-ETHANEDIOL, 2,1,3-benzothiadiazol-4-ylmethanamine, ACETATE ION, ... | | Authors: | Zak, K.M, Bostock, M.J, Ksiazek, M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Latency, thermal stability, and identification of an inhibitory compound of mirolysin, a secretory protease of the human periodontopathogen Tannerella forsythia .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7NP4
 
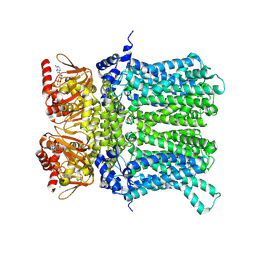 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
8VWL
 
 | |
7NWF
 
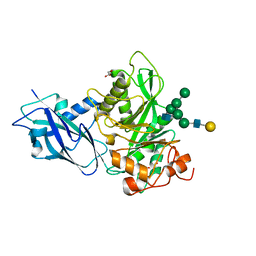 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with hybrid-type glycan (GalGlcNAcMan5GlcNAc) product | | Descriptor: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Garcia-Alija, M, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GH18 endo-beta-N-acetylglucosaminidases use distinct mechanisms to process hybrid-type N-linked glycans.
J.Biol.Chem., 297, 2021
|
|
7NP3
 
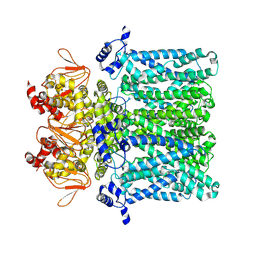 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2021-02-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
6YBV
 
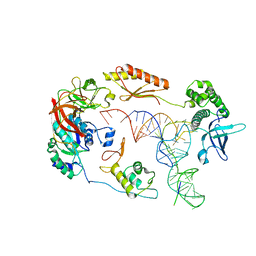 | | Structure of a human 48S translational initiation complex - eIF2-TC | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
7OFV
 
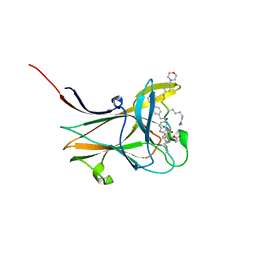 | | NMR-guided design of potent and selective EphA4 agonistic ligands | | Descriptor: | ACETATE ION, EphA4 agonist ligand, Ephrin type-A receptor 4 | | Authors: | Ganichkin, O.M, Craig, T.K, Baggio, C, Pellecchia, M. | | Deposit date: | 2021-05-05 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | NMR-Guided Design of Potent and Selective EphA4 Agonistic Ligands.
J.Med.Chem., 64, 2021
|
|
7VYW
 
 | |
8VA3
 
 | | Crystal structure of CapGH3b enzyme retrieved from capybara gut metagenome | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Martins, M.P, Morais, M.A.B, Chinaglia, M, Mandelli, F, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
8VWN
 
 | |
8VA4
 
 | | Crystal structure of CapGH16_3 enzyme retrieved from capybara gut metagenome | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 16, PHOSPHATE ION | | Authors: | Vieira, P.S, Martins, M.P, Morais, M.A.B, Mandelli, F, Chinaglia, M, Lima, E.A, Murakami, M.T. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A functionally augmented carbohydrate utilization locus from herbivore gut microbiota fueled by dietary beta-glucans.
NPJ Biofilms Microbiomes, 10, 2024
|
|
8WT8
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate) | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8WT7
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
