2POY
 
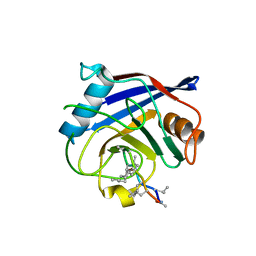 | | Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 in complex with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryptosporidium Parvum Cyclophilin Type Peptidyl-Prolyl Cis-Trans Isomerase Cgd2_4120 in Complex with Cyclosporin A.
To be Published
|
|
4V1O
 
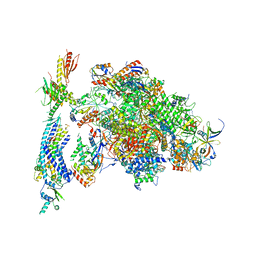 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
6ZCF
 
 | |
5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
3ZXZ
 
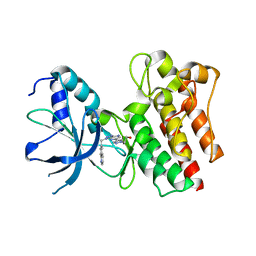 | | X-ray Structure of PF-04217903 bound to the kinase domain of c-Met | | Descriptor: | 2-{4-[1-(QUINOLIN-6-YLMETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRAZIN-6-YL]-1H-PYRAZOL-1-YL}ETHANOL, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-16 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
6ZCG
 
 | |
6ZCH
 
 | |
2PLU
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120 | | Descriptor: | 20k cyclophilin, putative | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_4120.
To be Published
|
|
8JJE
 
 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
3BF2
 
 | | Crystal structure of the A1KSW9_NEIMF protein from Neisseria meningitidis. Northeast Structural Genomics Consortium target MR36a | | Descriptor: | Putative lipoprotein | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Cunningham, K, Maglaqui, M, Owens, L, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the A1KSW9_NEIMF protein from Neisseria meningitidis.
To be Published
|
|
1HGV
 
 | | Filamentous Bacteriophage PH75 | | Descriptor: | PH75 INOVIRUS MAJOR COAT PROTEIN | | Authors: | Pederson, D.M, Welsh, L.C, Marvin, D.A, Sampson, M, Perham, R.N, Yu, M, Slater, M.R. | | Deposit date: | 2000-12-15 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | FIBER DIFFRACTION (2.4 Å) | | Cite: | The Protein Capsid of Filamentous Bacteriophage Ph75 from Thermus Thermophilus
J.Mol.Biol., 309, 2001
|
|
1OSC
 
 | | Crystal structure of rat CUTA1 at 2.15 A resolution | | Descriptor: | similar to divalent cation tolerant protein CUTA | | Authors: | Arnesano, F, Banci, L, Benvenuti, M, Bertini, I, Calderone, V, Mangani, S, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins
Suggests a Role in Signal Transduction
J.Biol.Chem., 278, 2003
|
|
3BFU
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3BGM
 
 | | Crystal Structure of PKD2 Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
2Q3X
 
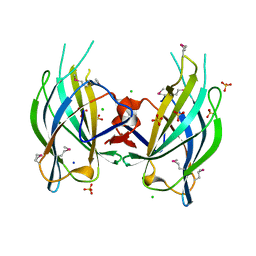 | | The RIM1alpha C2B domain | | Descriptor: | CHLORIDE ION, Regulating synaptic membrane exocytosis protein 1, SODIUM ION, ... | | Authors: | Guan, R, Dai, H, Tomchick, D.R, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2007-05-30 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the RIM1alpha C(2)B Domain at 1.7 A Resolution.
Biochemistry, 46, 2007
|
|
4V1N
 
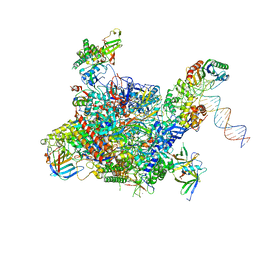 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
2A1B
 
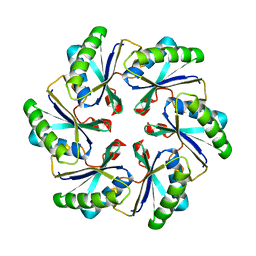 | | Carboxysome shell protein ccmK2 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 2 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-20 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2PYL
 
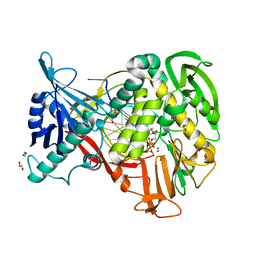 | | Phi29 DNA polymerase complexed with primer-template DNA and incoming nucleotide substrates (ternary complex) | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(CTGACGAATGTACA)-3', 5'-d(GACTGCTTAC(2DA))-3', ... | | Authors: | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2007-05-16 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
2A91
 
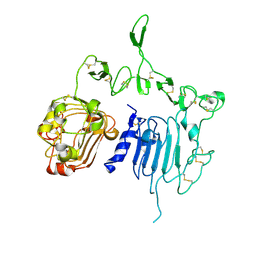 | | Crystal structure of ErbB2 domains 1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Garrett, T.P.J, McKern, N.M, Lou, M, Elleman, T.C, Adams, T.E, Lovrecz, G.O, Kofler, M, Jorissen, R.N, Nice, E.C, Burgess, A.W. | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of a Truncated ErbB2 Ectodomain Reveals an Active Conformation, Poised to Interact with Other ErbB Receptors
Mol.Cell, 11, 2003
|
|
1HF4
 
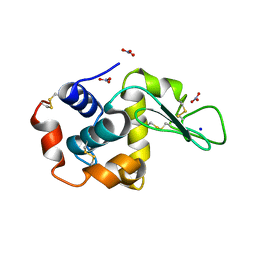 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1H4V
 
 | |
2AC2
 
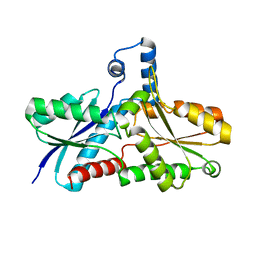 | | Crystal structure of the Tyr13Phe mutant variant of Bacillus subtilis Ferrochelatase with Zn(2+) bound at the active site | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2Q5R
 
 | | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase | | Descriptor: | Tagatose-6-phosphate kinase | | Authors: | McGrath, T.E, Soloveychik, M, Romanov, V, Thambipillai, D, Dharamsi, A, Virag, C, Domagala, M, Pai, E.F, Edwards, A.M, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2007-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo Staphylococcus aureus D-tagatose-6-phosphate kinase
TO BE PUBLISHED
|
|
3BU8
 
 | | Crystal Structure of TRF2 TRFH domain and TIN2 peptide complex | | Descriptor: | TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
3BKP
 
 | | Crystal structure of the Toxoplasma gondii cyclophilin, 49.m03261 | | Descriptor: | Cyclophilin, GLYCEROL | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Toxoplasma gondii cyclophilin, 49.m03261.
To be Published
|
|
