6Q69
 
 | |
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
7XN1
 
 | | Crystal structure of human acetylcholinesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase, ... | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
6Q6J
 
 | | Human phosphoserine phosphatase with substrate analogue homo-cysteic acid | | Descriptor: | (2~{S})-2-azanyl-4-sulfo-butanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wouters, J, Haufroid, M, Mirgaux, M. | | Deposit date: | 2018-12-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Crystal structures and snapshots along the reaction pathway of human phosphoserine phosphatase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QBR
 
 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200D, S203D | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Guillien, M, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S200D, S203D
To Be Published
|
|
6QE5
 
 | | Structure of E.coli RlmJ in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
6QF2
 
 | | X-Ray structure of Thermolysin crystallized on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QAB
 
 | | Human Butyrylcholinesterase in complex with (S)-N-(1-((2-cycloheptylethyl)amino)-3-(1H-indol-3-yl)-1-oxopropan-2-yl)-N,N-dimethylbutan-1-aminium | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Nachon, F, Harst, M, Knez, D, Gobec, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Tryptophan-derived butyrylcholinesterase inhibitors as promising leads against Alzheimer's disease.
Chem.Commun.(Camb.), 55, 2019
|
|
8RU5
 
 | | ATPase family AAA domain containing 2 with crystallization epitope mutations V1022R:Q1027E | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2 | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
6QJ4
 
 | | Crystal structure of the C. thermophilum condensin Ycs4-Brn1 subcomplex bound to the Smc4 ATPase head in complex with the C-terminal domain of Brn1 | | Descriptor: | Brn1, Condensin complex subunit 1,Condensin complex subunit 1,Condensin complex subunit 1, Condensin complex subunit 2, ... | | Authors: | Hassler, M, Haering, C.H, Kschonsak, M. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
6QD5
 
 | | X-ray Structure of the Human Urea Channel SLC14A1/UT1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, TETRAETHYLENE GLYCOL, Urea transporter 1, ... | | Authors: | Dietz, L, Chi, G, Pike, A.C.W, Moreau, C, Man, H, Snee, M, Scacioc, A, Shrestha, L, Mukhopadhyay, S.M.M, Mckinley, G, Ellis, K, Kliszcak, M, Chalk, R, Borkowska, O, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Durr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | X-ray Structure of the Human Urea Channel SLC14A1/UT1
To Be Published
|
|
8RU1
 
 | | Chromatin remodeling regulator CECR2 with in crystallo disulfide bond | | Descriptor: | Chromatin remodeling regulator CECR2, GLYCEROL, SODIUM ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
6QGS
 
 | | Crystal structure of APT1 bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, CHLORIDE ION, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
8RXR
 
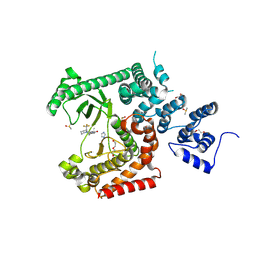 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
4WV8
 
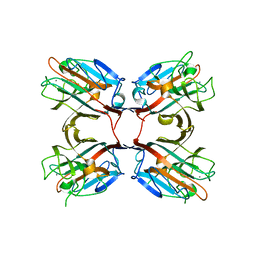 | | Crystal structure of a recombinant Vatairea macrocarpa seed lectin complexed with lactose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Seed lectin, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Cunha, R.M.S, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a Vatairea macrocarpa lectin in complex with a tumor-associated antigen: A new tool for cancer research.
Int.J.Biochem.Cell Biol., 72, 2016
|
|
4X8K
 
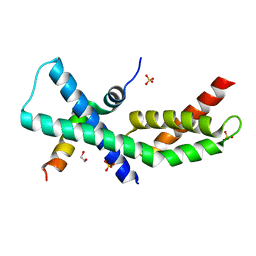 | | Mycobacterium tuberculosis RbpA-SID in complex with SigmaA domain 2 | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor SigA, RNA polymerase-binding protein RbpA, ... | | Authors: | Hubin, E.A, Flack, J.E, Tabib-Salazar, A, Paget, M.S, Darst, S.A, Campbell, E.A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6QE0
 
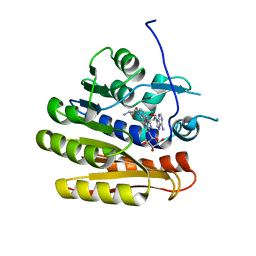 | | Structure of E.coli RlmJ in complex with a bisubstrate analogue (BA2) | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[2-[[9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]ethyl]amino]-2-azanyl-butanoic acid, Ribosomal RNA large subunit methyltransferase J | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-03 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
6Q67
 
 | |
7QFB
 
 | | Crystal structure of Protein Phosphatase 1 in complex with PP1-binding peptide from PTG | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Protein phosphatase 1 regulatory subunit 3C, ... | | Authors: | Semrau, M.S, Storici, P, Lolli, G. | | Deposit date: | 2021-12-05 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun, 13, 2022
|
|
6QF3
 
 | | X-Ray structure of Thermolysin soaked with sodium aspartate on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ASPARTIC ACID, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
8G2D
 
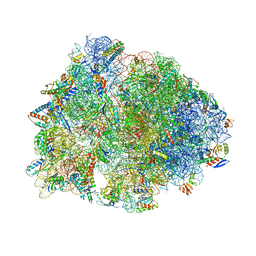 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
7QF7
 
 | |
8VQ2
 
 | | HSV1 polymerase ternary complex with dsDNA and compound 44 | | Descriptor: | 2-(4-bromophenyl)-N-(3-methoxy-4-{[(4S)-2-oxo-1,3-oxazolidin-4-yl]methyl}phenyl)acetamide, DNA (5'-D(P*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Hayes, R.P, Heo, M.R, Plotkin, M. | | Deposit date: | 2024-01-17 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.829 Å) | | Cite: | Discovery of Broad-Spectrum Herpes Antiviral Oxazolidinone Amide Derivatives and Their Structure-Activity Relationships.
Acs Med.Chem.Lett., 15, 2024
|
|
9FQA
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor PSB-21101 (compound 30b in publication) | | Descriptor: | (5-chloranylpyridin-3-yl) 2-fluoranyl-4-phenylmethoxy-benzoate, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Oneto, A, Guetschow, M, Mueller, C.E. | | Deposit date: | 2024-06-14 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Nonpeptidic Irreversible Inhibitors of SARS-CoV-2 Main Protease with Potent Antiviral Activity.
J.Med.Chem., 2024
|
|
9FQ9
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor PSB-21110 (compound 29b in publication) | | Descriptor: | (5-chloranylpyridin-3-yl) 4-ethoxy-2-fluoranyl-benzoate, BROMIDE ION, Non-structural protein 11 | | Authors: | Strater, N, Claff, T, Sylvester, K, Oneto, A, Guetschow, M, Mueller, C.E. | | Deposit date: | 2024-06-19 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Nonpeptidic Irreversible Inhibitors of SARS-CoV-2 Main Protease with Potent Antiviral Activity.
J.Med.Chem., 2024
|
|
