1UZM
 
 | | MabA from Mycobacterium tuberculosis | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE, CESIUM ION | | Authors: | Cohen-Gonsaud, M, Ducasse, S, Quemard, A, Labesse, G. | | Deposit date: | 2004-03-14 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of Maba from Mycobacterium Tuberculosis, a Reductase Involved in Long-Chain Fatty Acid Biosynthesis.
J.Mol.Biol., 320, 2002
|
|
1V2E
 
 | |
1V3I
 
 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
1V0B
 
 | | Crystal structure of the t198a mutant of pfpk5 | | Descriptor: | CELL DIVISION CONTROL PROTEIN 2 HOMOLOG | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2004-03-26 | | Release date: | 2004-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
1V5T
 
 | | Solution Structure of the Ubiquitin-like Domain from Mouse Hypothetical 8430435I17Rik Protein | | Descriptor: | 8430435I17Rik Protein | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-like Domain from Mouse Hypothetical 8430435I17Rik Protein
To be Published
|
|
1H85
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH VAL 136 REPLACED BY LEU (V136L) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a Cluster of Hydrophobic Residues Near the Fad Cofactor in Anabaena Pcc 7119 Ferredoxin-Nadp+ Reductase for Optimal Complex Formation and Electron Transfer to Ferredoxin
J.Biol.Chem., 276, 2001
|
|
1V5L
 
 | | Solution structure of PDZ domain of mouse Alpha-actinin-2 associated LIM protein | | Descriptor: | PDZ and LIM domain 3; actinin alpha 2 associated LIM protein | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PDZ domain of mouse Alpha-actinin-2 associated LIM protein
To be Published
|
|
1V86
 
 | | Solution structure of the ubiquitin domain from mouse D7Wsu128e protein | | Descriptor: | DNA segment, Chr 7, Wayne State University 128, ... | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin domain from mouse D7Wsu128e protein
To be Published
|
|
1VAU
 
 | | Xenon derivative of hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Takeda, K, Miyatake, H, Park, S.Y, Kawamoto, M, Kamiya, N, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-19 | | Release date: | 2005-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multi-wavelength anomalous diffraction method for I and Xe atoms using ultra-high-energy X-rays from SPring-8
J.Appl.Crystallogr., 37, 2004
|
|
1GTV
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH THYMIDINE-5'-DIPHOSPHATE (TDP) | | Descriptor: | ACETATE ION, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Ursby, T, Weik, M, Fioravanti, E, Delarue, M, Goeldner, M, Bourgeois, D. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cryophotolysis of Caged Compounds: A Technique for Trapping Intermediate States in Protein Crystals
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1UNG
 
 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | 6-PHENYL[5H]PYRROLO[2,3-B]PYRAZINE, CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNH
 
 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | Authors: | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | Deposit date: | 2003-09-10 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UPR
 
 | | Crystal structure of the PEPP1 pleckstrin homology domain in complex with Inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY A MEMBER 4 | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
1URH
 
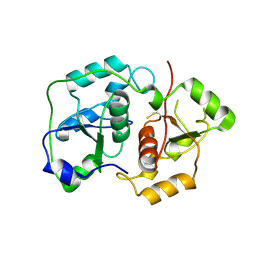 | | The "Rhodanese" fold and catalytic mechanism of 3-mercaptopyruvate sulfotransferases: Crystal structure of SseA from Escherichia coli | | Descriptor: | 3-MERCAPTOPYRUVATE SULFURTRANSFERASE, SULFITE ION | | Authors: | Spallarossa, A, Forlani, F, Carpen, A, Armirotti, A, Pagani, S, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-10-30 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The "Rhodanese" Fold and Catalytic Mechanism of 3-Mercaptopyruvate Sulfurtransferases: Crystal Structure of Ssea from Escherichia Coli
J.Mol.Biol., 335, 2004
|
|
1UX1
 
 | | Bacillus subtilis cytidine deaminase with a Cys53His and an Arg56Gln substitution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ... | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1UTT
 
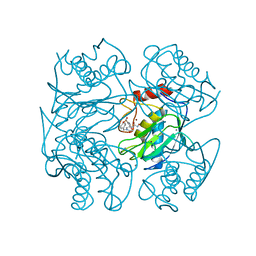 | | Crystal Structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL) ETHYL-4-(4'-ETHOXY [1,1'-BIPHENYL]-4-YL)-4-OXOBUTANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J. Mol. Biol., 341, 2004
|
|
1UWZ
 
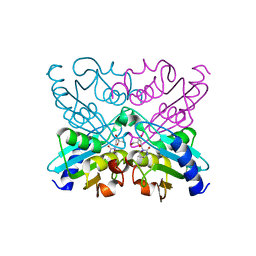 | | Bacillus subtilis cytidine deaminase with an Arg56 - Ala substitution | | Descriptor: | CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ZINC ION | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1UPL
 
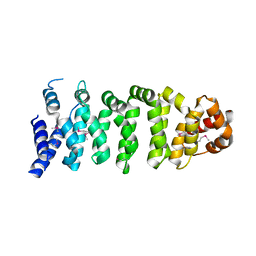 | | Crystal structure of MO25 alpha | | Descriptor: | MO25 PROTEIN | | Authors: | Milburn, C.C, Boudeau, J, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-07 | | Release date: | 2004-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mo25 Alpha in Complex with the C-Terminus of the Pseudo Kinase Ste-20 Related Adaptor (Strad)
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UTZ
 
 | | Crystal Structure of MMP-12 complexed to (2R)-3-({[4-[(pyridin-4-yl)phenyl]-thien-2-yl}carboxamido)(phenyl)propanoic acid | | Descriptor: | (2R)-3-({[4-[(PYRIDIN-4-YL)PHENYL]-THIEN-2-YL}CARBOXAMIDO)(PHENYL)PROPANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Novel Non-Peptidic, Non-Zinc Chelating Inhibitors Bound to Mmp-12
J.Mol.Biol., 341, 2004
|
|
1V08
 
 | | Crystal structure of the Zea maze beta-glucosidase-1 in complex with gluco-tetrazole | | Descriptor: | BETA-GLUCOSIDASE, NOJIRIMYCINE TETRAZOLE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1UOS
 
 | | The Crystal Structure of the Snake Venom Toxin Convulxin | | Descriptor: | CONVULXIN ALPHA, CONVULXIN BETA | | Authors: | Batuwangala, T, Leduc, M, Gibbins, J.M, Bon, C, Jones, E.Y. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Snake-Venom Toxin Convulxin
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U64
 
 | | The Solution Structure of d(G3T4G4)2 | | Descriptor: | 5'-D(*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3' | | Authors: | Sket, P, Crnugelj, M, Plavec, J. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or NH4+ ions.
Bioorg.Med.Chem., 12, 2004
|
|
1UBZ
 
 | |
1UF1
 
 | | Solution structure of the second PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 protein | | Authors: | Saito, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of human KIAA1526 protein
To be Published
|
|
1U0R
 
 | | Crystal structure of Mycobacterium tuberculosis NAD kinase | | Descriptor: | Inorganic polyphosphate/ATP-NAD kinase | | Authors: | Garavaglia, S, Raffaelli, N, Finaurini, L, Magni, G, Rizzi, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-14 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel fold revealed by Mycobacterium tuberculosis NAD kinase, a key allosteric enzyme in NADP biosynthesis
J.Biol.Chem., 279, 2004
|
|
