5JWD
 
 | | Crystal structure of H-2Db in complex with the LCMV-derived GP392-401 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
8POK
 
 | | Cryo-EM structure of cell-free synthesized human histamine H2 receptor coupled to heterotrimeric Gs protein in lipid environment | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HISTAMINE, ... | | Authors: | Schnelle, K, Koeck, Z, Persechino, M, Umbach, S, Schihada, H, Januliene, D, Parey, K, Pockes, S, Kolb, P, Doetsch, V, Moeller, A, Hilger, D, Bernhard, F. | | Deposit date: | 2023-07-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of cell-free synthesized human histamine 2 receptor/G s complex in nanodisc environment.
Nat Commun, 15, 2024
|
|
6YVL
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 1.42 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVM
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.13 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVN
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.84 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
8PP6
 
 | | human RYBP-PRC1 bound to H2AK118ub1 nucleosome | | Descriptor: | DNA (215-MER), Histone H2A, Histone H2B, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5K2Z
 
 | | PDX1.3-adduct (Arabidopsis) | | Descriptor: | 1,2-ETHANEDIOL, 2-azanylpenta-1,4-dien-3-one, CHLORIDE ION, ... | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | Deposit date: | 2016-05-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4IGR
 
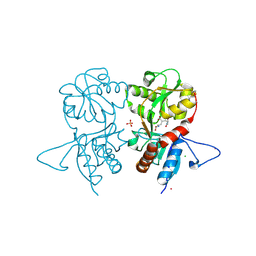 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist ZA302 | | Descriptor: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2012-12-18 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
5GO7
 
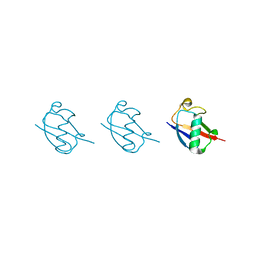 | | Linear tri-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
7RZC
 
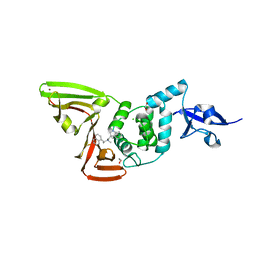 | | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor | | Descriptor: | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor
To be Published
|
|
5GOC
 
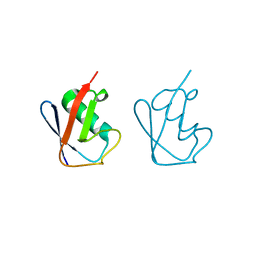 | | Lys11-linked diubiquitin | | Descriptor: | D-ubiquitin, SODIUM ION, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
6YWT
 
 | |
5GOI
 
 | | Lys48-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
3AVO
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with Pantothenate | | Descriptor: | CITRATE ANION, GLYCEROL, PANTOTHENOIC ACID, ... | | Authors: | Chetnani, B, Kumar, P, Abhinav, K.V, Chhibber, M, Surolia, A, Vijayan, M. | | Deposit date: | 2011-03-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Location and conformation of pantothenate and its derivatives in Mycobacterium tuberculosis pantothenate kinase: insights into enzyme action
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1IDP
 
 | |
6YVO
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 3.55 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
3AWE
 
 | | Crystal structure of Pten-like domain of Ci-VSP (248-576) | | Descriptor: | ACETIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Matsuda, M, Sakata, S, Takeshita, K, Suzuki, M, Yamashita, E, Okamura, Y, Nakagawa, A. | | Deposit date: | 2011-03-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity
J.Biol.Chem., 286, 2011
|
|
6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-30 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7TG5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the Presence of Fe Ion from Yersinia pestis | | Descriptor: | CHLORIDE ION, FE (III) ION, Pirin family protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the presence of Fe ion from Yersinia pestis
To Be Published
|
|
7TFQ
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, FORMIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis
To Be Published
|
|
3AU3
 
 | | Crystal structure of armadillo repeat domain of APC | | Descriptor: | Adenomatous polyposis coli protein | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-28 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
7THH
 
 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
7TH8
 
 | | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds | | Descriptor: | Nodule cysteine-rich protein 13 | | Authors: | Buchko, G.W, Zhou, M, Shah, D.M, Velivelli, S.L.S. | | Deposit date: | 2022-01-10 | | Release date: | 2022-02-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds
To Be Published
|
|
3AUJ
 
 | | Structure of diol dehydratase complexed with glycerol | | Descriptor: | CALCIUM ION, COBALAMIN, Diol dehydrase alpha subunit, ... | | Authors: | Yamanishi, M, Kinoshita, K, Fukuoka, M, Shibata, T, Tobimatsu, T, Toraya, T. | | Deposit date: | 2011-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redesign of coenzyme B(12) dependent diol dehydratase to be resistant to the mechanism-based inactivation by glycerol and act on longer chain 1,2-diols
Febs J., 279, 2012
|
|
