1D4F
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT RAT-LIVER D244E MUTANT S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Komoto, J, Huang, Y, Takusagawa, F, Gomi, T, Ogawa, H, Takata, Y, Fujioka, M. | | Deposit date: | 2000-06-22 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Effects of site-directed mutagenesis on structure and function of recombinant rat liver S-adenosylhomocysteine hydrolase. Crystal structure of D244E mutant enzyme.
J.Biol.Chem., 275, 2000
|
|
3NOY
 
 | | Crystal structure of IspG (gcpE) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Bacher, A. | | Deposit date: | 2010-06-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biosynthesis of isoprenoids: crystal structure of the [4Fe-4S] cluster protein IspG.
J.Mol.Biol., 404, 2010
|
|
5CHS
 
 | |
1T3K
 
 | | NMR structure of a CDC25-like dual-specificity tyrosine phosphatase of Arabidopsis thaliana | | Descriptor: | Dual-specificity tyrosine phosphatase, ZINC ION | | Authors: | Landrieu, I, da Costa, M, De Veylder, L, Dewitte, F, Vandepoele, K, Hassan, S, Wieruszeski, J.M, Faure, J.D, Inze, D, Lippens, G. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1D6B
 
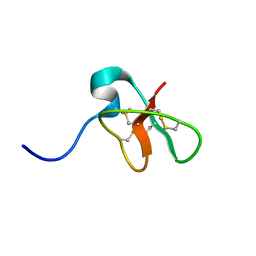 | | SOLUTION STRUCTURE OF DEFENSIN-LIKE PEPTIDE-2 (DLP-2) FROM PLATYPUS VENOM | | Descriptor: | DEFENSIN-LIKE PEPTIDE-2 | | Authors: | Torres, A.M, De Plater, G.M, Doverskog, M, C Birinyi-Strachan, L, Nicholson, G.M, Gallagher, C.H, Kuchel, P.W. | | Deposit date: | 1999-10-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Defensin-like peptide-2 from platypus venom: member of a class of peptides with a distinct structural fold.
Biochem.J., 348, 2000
|
|
1INH
 
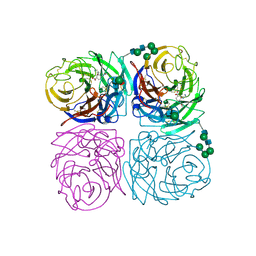 | | INFLUENZA A SUBTYPE N2 NEURAMINIDASE COMPLEXED WITH AROMATIC BANA111 INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(ACETYLAMINO)-3-[(AMINOACETYL)AMINO]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1995-07-07 | | Release date: | 1996-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitors of influenza virus sialidase. A benzoic acid lead with novel interaction.
J.Med.Chem., 38, 1995
|
|
4OAZ
 
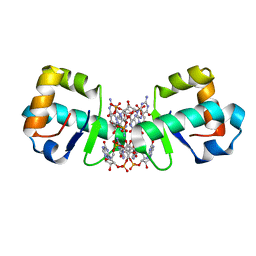 | | BldD CTD-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative DNA-binding protein | | Authors: | Schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R.G. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
1IRJ
 
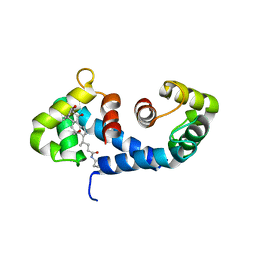 | | Crystal Structure of the MRP14 complexed with CHAPS | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, Migration Inhibitory Factor-Related Protein 14 | | Authors: | Itou, H, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2001-10-09 | | Release date: | 2002-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of human MRP14 (S100A9), a Ca(2+)-dependent regulator protein in inflammatory process.
J.Mol.Biol., 316, 2002
|
|
4OB3
 
 | | Crystal Structure of Nitrile Hydratase from Pseudonocardia thermophila : A Reference Structure to Boronic Acid Inhibition of Nitrile Hydratase | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta, ... | | Authors: | Rui, W, Salette, M, Ruslan, S, Richard, H, Dali, L. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-26 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The active site sulfenic acid ligand in nitrile hydratases can function as a nucleophile.
J.Am.Chem.Soc., 136, 2014
|
|
4OC0
 
 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CCIBzL, a urea-based inhibitor N~2~-[(1-carboxycyclopropyl)carbamoyl]-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5CZ8
 
 | |
5D0Z
 
 | |
2Q0Q
 
 | | Structure of the Native M. Smegmatis Aryl Esterase | | Descriptor: | GLYCEROL, SULFATE ION, aryl esterase | | Authors: | Mathews, I.I, Soltis, M, Saldajeno, M, Ganshaw, G, Sala, R, Weyler, W, Cervin, M.A, Whited, G, Bott, R. | | Deposit date: | 2007-05-22 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a novel enzyme that catalyzes acyl transfer to alcohols in aqueous conditions.
Biochemistry, 46, 2007
|
|
2Q29
 
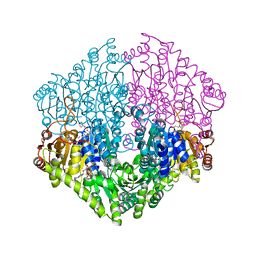 | | Crystal structure of oxalyl-coA decarboxylase from Escherichia coli in complex with acetyl coenzyme A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Werther, T, Zimmer, A, Wille, G, Hubner, G, Weiss, M.S, Konig, S. | | Deposit date: | 2007-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | New insights into structure-function relationships of oxalyl CoA decarboxylase from Escherichia coli.
Febs J., 277, 2010
|
|
1J1C
 
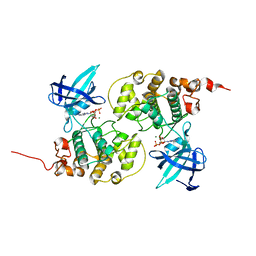 | | Binary complex structure of human tau protein kinase I with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase kinase-3 beta, MAGNESIUM ION | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TJP
 
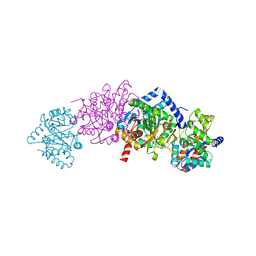 | | Crystal Structure Of Wild-Type Tryptophan Synthase Complexed With 1-[(2-hydroxylphenyl)amino]3-glycerolphosphate | | Descriptor: | 1-[(2-HYDROXYLPHENYL)AMINO]3-GLYCEROLPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2004-06-07 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
1H3L
 
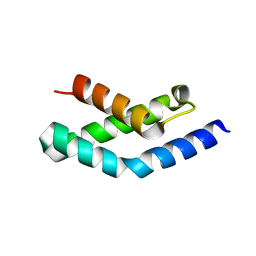 | | N-terminal fragment of SigR from Streptomyces coelicolor | | Descriptor: | RNA POLYMERASE SIGMA FACTOR | | Authors: | Li, W, Stevenson, C.E.M, Burton, N, Jakimowicz, P, Paget, M.S.B, Buttner, M.J, Lawson, D.M, Kleanthous, C. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Identification and Structure of the Anti-Sigma Factor-Binding Domain of the Disulfide-Stress Regulated Sigma Factor Sigma(R) from Streptomyces Coelicolor
J.Mol.Biol., 323, 2002
|
|
5D0T
 
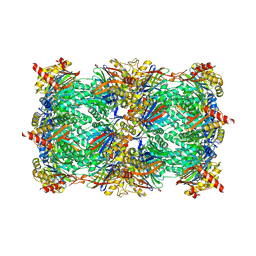 | | Yeast 20S proteasome beta5-D166N mutant in complex with MG132 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
1Z9C
 
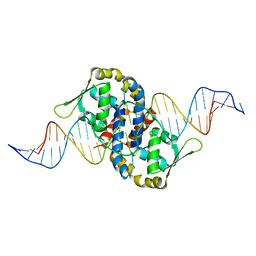 | | Crystal structure of OhrR bound to the ohrA promoter: Structure of MarR family protein with operator DNA | | Descriptor: | DNA (29-MER), Organic hydroperoxide resistance transcriptional regulator | | Authors: | Hong, M, Fuangthong, M, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2005-04-01 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of an OhrR-ohrA Operator Complex Reveals the DNA Binding Mechanism of the MarR Family.
Mol.Cell, 20, 2005
|
|
7XSB
 
 | |
1MOO
 
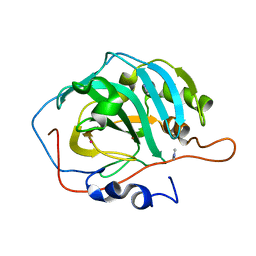 | | Site Specific Mutant (H64A) of Human Carbonic Anhydrase II at high resolution | | Descriptor: | 4-METHYLIMIDAZOLE, Carbonic Anhydrase II, MERCURY (II) ION, ... | | Authors: | Duda, D.M, Govindasamy, L, Agbandje-McKenna, M, Tu, C.K, Silverman, D.N, McKenna, R. | | Deposit date: | 2002-09-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The refined atomic structure of carbonic anhydrase II at 1.05 A resolution: implications of chemical rescue of proton transfer.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1Z3J
 
 | | Solution Structure of MMP12 in the presence of N-isobutyl-N-4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.M, Luchinat, C, Mangani, S, Terni, B, Turano, P. | | Deposit date: | 2005-03-13 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1UUR
 
 | | Structure of an activated Dictyostelium STAT in its DNA-unbound form | | Descriptor: | STATA PROTEIN | | Authors: | Soler-Lopez, M, Petosa, C, Fukuzawa, M, Ravelli, R, Williams, J.G, Muller, C.W. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an Activated Dictyostelium Stat in its DNA-Unbound Form
Mol.Cell, 13, 2004
|
|
1UYK
 
 | | Human Hsp90-alpha with 8-Benzo[1,3]dioxol-,5-ylmethyl-9-butyl-2-fluoro-9H-purin-6-ylamine | | Descriptor: | 8-BENZO[1,3]DIOXOL-,5-YLMETHYL-9-BUTYL-2-FLUORO-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1Z91
 
 | |
