8QNF
 
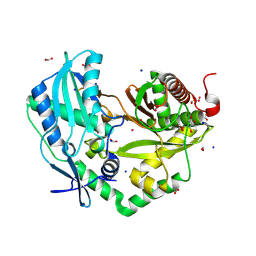 | | Crystal structure of the Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase, FORMIC ACID, GLYCEROL, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
1ZZ2
 
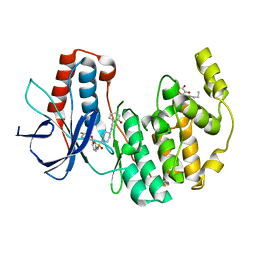 | | Two Classes of p38alpha MAP Kinase Inhibitors Having a Common Diphenylether Core but Exhibiting Divergent Binding Modes | | Descriptor: | Mitogen-activated protein kinase 14, N-[3-(4-FLUOROPHENOXY)PHENYL]-4-[(2-HYDROXYBENZYL)AMINO]PIPERIDINE-1-SULFONAMIDE, octyl beta-D-glucopyranoside | | Authors: | Michelotti, E.L, Moffett, K.K, Springman, E.B, Karpusas, M. | | Deposit date: | 2005-06-13 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two classes of p38alpha MAP kinase inhibitors having a common diphenylether core but exhibiting divergent binding modes.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6ULM
 
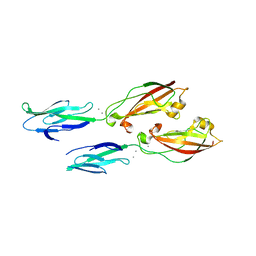 | | Crystal structure of human cadherin 17 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-17 | | Authors: | Gray, M.E, Sotomayor, M. | | Deposit date: | 2019-10-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the nonclassical cadherin-17 N-terminus and implications for its adhesive binding mechanism.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6UD0
 
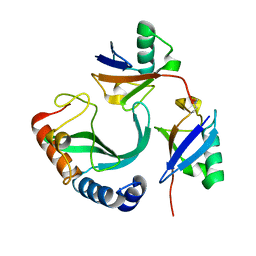 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
7TLX
 
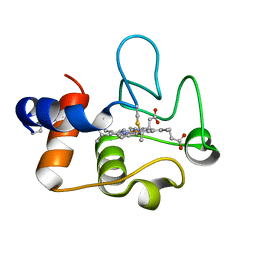 | | Crystal Structure of cytochrome c from Pseudomonas putida S16 | | Descriptor: | C-type cytochrome, HEME C | | Authors: | Wu, K, Dulchavsky, M, Stull, F, Bardwell, J.C.A. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The enzyme pseudooxynicotine amine oxidase from Pseudomonas putida S16 is not an oxidase, but a dehydrogenase.
J.Biol.Chem., 298, 2022
|
|
5AXN
 
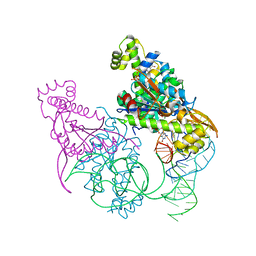 | | Crystal structure of Thg1 like protein (TLP) with tRNA(Phe) and GDPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RNA (75-MER), ... | | Authors: | Kimura, S, Suzuki, T, Yu, J, Kato, K, Yao, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Template-dependent nucleotide addition in the reverse (3'-5') direction by Thg1-like protein
Sci Adv, 2, 2016
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
5AV6
 
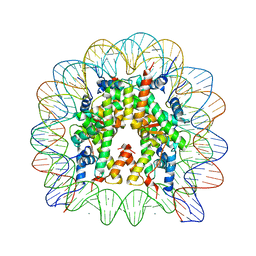 | | human nucleosome core particle | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Wakamori, M, Fujii, Y, Umehara, T, Yokoyama, S. | | Deposit date: | 2015-06-12 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation
Sci Rep, 5, 2015
|
|
6NZS
 
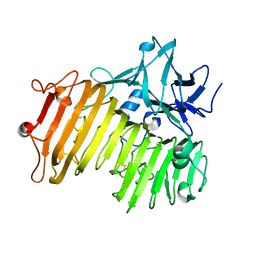 | | Dextranase AoDex KQ11 | | Descriptor: | Dextranase | | Authors: | Ren, W, Yan, W, Gu, L, Feng, Y, Dong, D, Wang, S, Wang, C, Lyu, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
To Be Published
|
|
1RZG
 
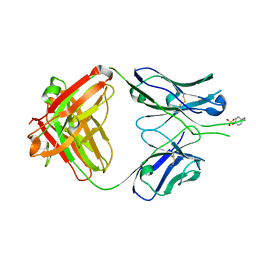 | | Crystal structure of Human anti-HIV-1 GP120 reactive antibody 412d | | Descriptor: | ASPARTIC ACID, CYSTEINE, Fab 412d heavy chain, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6QO8
 
 | |
5AZX
 
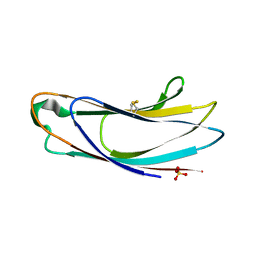 | | Crystal structure of p24delta1 GOLD domain (native 1) | | Descriptor: | SULFATE ION, Transmembrane emp24 domain-containing protein 10 | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2015-10-23 | | Release date: | 2016-09-14 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 3D Structure and Interaction of p24 beta and p24 delta Golgi Dynamics Domains: Implication for p24 Complex Formation and Cargo Transport
J.Mol.Biol., 428, 2016
|
|
6O0X
 
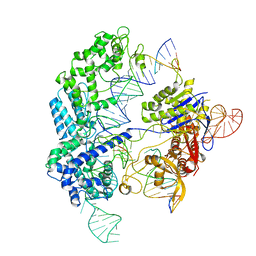 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
5B1I
 
 | |
7VE3
 
 | | Structure of the complex of sheep lactoperoxidase with hypoiodite at 2.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, P.K, Yamini, S, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence of the oxidation of iodide ion into hyper-reactive hypoiodite ion by mammalian heme lactoperoxidase.
Protein Sci., 31, 2022
|
|
5B2N
 
 | | Crystal structure of the light-driven chloride ion-pumping rhodopsin, ClP, from Nonlabens marinus | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, DECANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2016-01-20 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structural Mechanism for Light-driven Transport by a New Type of Chloride Ion Pump, Nonlabens marinus Rhodopsin-3
J.Biol.Chem., 291, 2016
|
|
6MYY
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 3 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6MZQ
 
 | | TAS-120 in reversible binding mode with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1 | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
6MZW
 
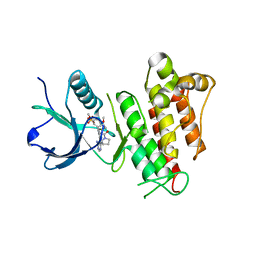 | | TAS-120 covalent complex with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
6QMP
 
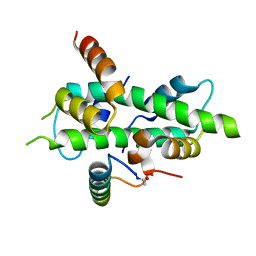 | | NF-YB/C Heterodimer in Complex with NF-YA Peptide | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
1LO2
 
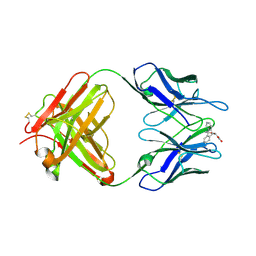 | | Retro-Diels-Alderase Catalytic Antibody | | Descriptor: | If kappa light chain, Ig gamma 2a heavy chain, [2'-CARBOXYLETHYL]-10-METHYL-ANTHRACENE ENDOPEROXIDE | | Authors: | Hugot, M, Reymond, J.L, Baumann, U. | | Deposit date: | 2002-05-06 | | Release date: | 2002-06-26 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for the activity of retro-Diels-Alder catalytic antibodies: evidence for a catalytic aromatic residue.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1D78
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
6JZU
 
 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6QNS
 
 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B mutant I1248W/V1249W in complex with human synaptotagmin 1 and GD1a receptors | | Descriptor: | Botulinum neurotoxin type B, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Synaptotagmin-1 | | Authors: | Masuyer, G, Yin, L, Zhang, S, Miyashita, S.I, Dong, M, Stenmark, P. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a membrane binding loop leads to engineering botulinum neurotoxin B with improved therapeutic efficacy.
Plos Biol., 18, 2020
|
|
