5OV7
 
 | | tubulin - rigosertib complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Menchon, G, Prota, A.E, Steinmetz, M, Jost, M. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent.
Mol. Cell, 68, 2017
|
|
6U7L
 
 | | 2.75 Angstrom Crystal Structure of Galactarate Dehydratase from Escherichia coli. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactarate dehydratase (L-threo-forming) | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment.
Protein Sci., 29, 2020
|
|
8JCT
 
 | |
8J09
 
 | | Crystal structure of protein 3745 | | Descriptor: | Cell division control protein 45, DNA replication regulator SLD3 | | Authors: | Li, H, Yao, M. | | Deposit date: | 2023-04-10 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into a process of complex formation by 3745
To Be Published
|
|
4MQE
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor in the apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4MV5
 
 | | IspH in complex with 6-chloropyridin-3-ylmethyl diphosphate | | Descriptor: | (6-chloropyridin-3-yl)methyl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
5WQP
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WST
 
 | | Crystal structure of Myo7a SAH | | Descriptor: | Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
5WW0
 
 | | Crystal structure of Set7, a novel histone methyltransferase in Schizossacharomyces pombe | | Descriptor: | SET domain-containing protein 7, SULFATE ION | | Authors: | Mevius, D.E.H.F, Shen, Y, Morishita, M, Carrozzini, B, Caliandro, R, di Luccio, E. | | Deposit date: | 2016-12-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis.
Structure, 27, 2019
|
|
4N1J
 
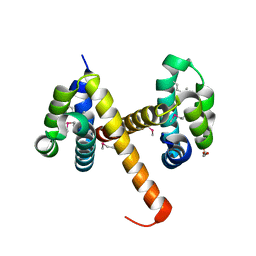 | | Crystal structures of NLRP14 pyrin domain reveal a conformational switch mechanism, regulating its molecular interactions | | Descriptor: | GLYCEROL, NACHT, LRR and PYD domains-containing protein 14 | | Authors: | Eibl, C, Hessenberger, M, Wenger, J, Brandstetter, H. | | Deposit date: | 2013-10-04 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N2E
 
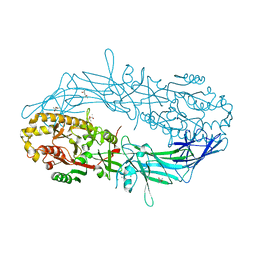 | | Crystal structure of Protein Arginine Deiminase 2 (D123N, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2B
 
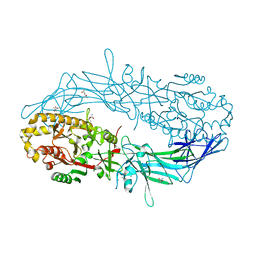 | | Crystal structure of Protein Arginine Deiminase 2 (10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5WOH
 
 | |
8K7X
 
 | | Crystal structure of GH146 beta-L-arabinofuranosidase Bll3HypBA1 (amino acids 380-1223) in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pan, L, Maruyama, S, Miyake, M, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-07-27 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bifidobacterial GH146 beta-L-arabinofuranosidase for the removal of beta 1,3-L-arabinofuranosides on plant glycans.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
5OW0
 
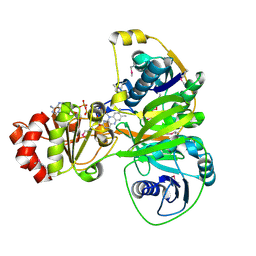 | | Crystal structure of an electron transfer flavoprotein from Geobacter metallireducens | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein, alpha subunit, ... | | Authors: | Essen, L.-O, Vogt, M.S, Heider, J, Koelzer, S, Peschke, P, Chowdhury, N.P, Schuehle, K, Kleinsorge, D. | | Deposit date: | 2017-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of an electron transfer flavoprotein involved in toluene degradation in strictly anaerobic bacteria.
J.Bacteriol., 2019
|
|
5WP1
 
 | | Complex of ERK2 with 5,7-dihydroxychromone | | Descriptor: | 5,7-dihydroxy-4H-1-benzopyran-4-one, BENZOIC ACID, Mitogen-activated protein kinase 1, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiple phytochemicals at low doses accumulatively inhibit one key protein in cancers
To Be Published
|
|
4N7M
 
 | |
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WUE
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
4MJI
 
 | | T cell response to a HIV reverse transcriptase epitope presented by the protective allele HLA-B*51:01 | | Descriptor: | Beta-2-microglobulin, HIV Reverse Transcriptase peptide Marker, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Motozono, C, Takiguchi, M. | | Deposit date: | 2013-09-03 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01
J.Immunol., 192, 2014
|
|
8MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*UP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-06 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
4MNR
 
 | | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta
To be Published
|
|
8ODQ
 
 | | SufS-SufU complex from Mycobacterium tuberculosis | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Cysteine desulfurase, NITRATE ION, ... | | Authors: | Elchennawi, I, Carpentier, P, Caux, C, Ponge, M, Ollagnier de Choudens, S. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium tuberculosis Zinc SufU-SufS Complex.
Biomolecules, 13, 2023
|
|
5WX8
 
 | | Human herpesvirus 6A immediate early protein 2 C-terminal domain | | Descriptor: | Immediate-early protein 2 | | Authors: | Nishimura, M, Wang, J, Wakata, A, Sakamoto, K, Mori, Y. | | Deposit date: | 2017-01-06 | | Release date: | 2017-08-02 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of Human Herpesvirus 6A Immediate Early Protein 2.
J. Virol., 91, 2017
|
|
4MR7
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP54626 | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
