6EPA
 
 | | Structure of dNCS-1 bound to the NCS-1/Ric8a protein/protein interaction regulator IGS-1.76 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, FI18190p1, ... | | Authors: | Sanchez-Barrena, M.J, Daniel, M, Infantes, L. | | Deposit date: | 2017-10-11 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Deciphering the Inhibition of the Neuronal Calcium Sensor 1 and the Guanine Exchange Factor Ric8a with a Small Phenothiazine Molecule for the Rational Generation of Therapeutic Synapse Function Regulators.
J. Med. Chem., 61, 2018
|
|
3BYR
 
 | | Mode of Action of a Putative Zinc Transporter CzrB (Zn form) | | Descriptor: | CzrB protein, ZINC ION | | Authors: | Cherezov, V, Srinivasan, V, Szebenyi, D.M.E, Caffrey, M. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the Mode of Action of a Putative Zinc Transporter CzrB in Thermus thermophilus
Structure, 16, 2008
|
|
3BZZ
 
 | | Crystal structural of the mutated R313T EscU/SpaS C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.407 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
1QX5
 
 | |
1QXB
 
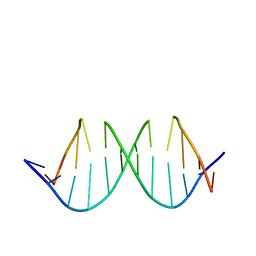 | | NMR structure determination of the self complementary DNA Dodecamer CGCGAATT*CGCG in which a ribose is inserted between the 3'-OH of T8 and the 5'-phosphate group of C9 | | Descriptor: | 5'-d(CpGpCpGpApApTpTpCpGpCpG)-3', beta-D-ribofuranose | | Authors: | Nauwelaerts, K, Vastmans, K, Froeyen, M, Kempeneers, V, Rozenski, J, Rosemeyer, H, Van Aerschot, A, Busson, R, Efimtseva, E, Mikhailov, S, Lescrinier, E, Herdewijn, P. | | Deposit date: | 2003-09-05 | | Release date: | 2004-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cleavage of DNA without loss of genetic information by incorporation of a disaccharide nucleoside.
Nucleic Acids Res., 31, 2003
|
|
2Q6V
 
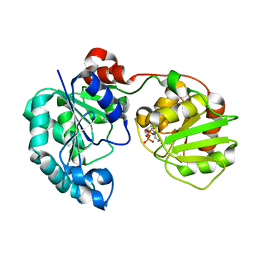 | | Crystal Structure of GumK in complex with UDP | | Descriptor: | Glucuronosyltransferase GumK, URIDINE-5'-DIPHOSPHATE | | Authors: | Barreras, M. | | Deposit date: | 2007-06-05 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure and mechanism of GumK, a membrane-associated glucuronosyltransferase.
J.Biol.Chem., 283, 2008
|
|
1QY7
 
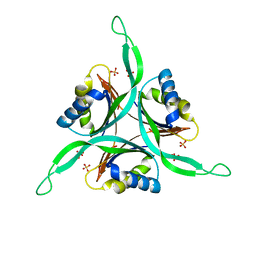 | | The structure of the PII protein from the cyanobacteria Synechococcus sp. PCC 7942 | | Descriptor: | NICKEL (II) ION, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Clancy, P, Garcia-Dominguez, M, Forchhammer, K, Florencio, F, Tandeau de Marsac, N, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 2003-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of the PII proteins from the cyanobacteria Synechococcus sp. PCC 7942 and Synechocystis sp. PCC 6803.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2Q8L
 
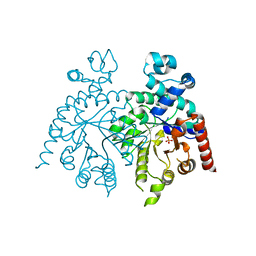 | | Crystal structure of orotidine 5'-phosphate decarboxylase from Plasmodium falciparum | | Descriptor: | Orotidine-monophosphate-decarboxylase, PHOSPHATE ION | | Authors: | Liu, Y, Lau, W, Lew, J, Amani, M, Hui, R, Pai, E.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of orotidine 5'-phosphate decarboxylase from Plasmodium falciparum.
To be Published
|
|
3C5F
 
 | | Structure of a binary complex of the R517A Pol lambda mutant | | Descriptor: | DNA (5'-D(*DCP*DAP*DGP*DTP*DAP*DC)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DCP*DGP*DTP*DAP*DCP*DTP*DG)-3'), DNA (5'-D(P*DGP*DCP*DCP*DG)-3'), ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Foley, M.C, Pedersen, L.C, Schlick, T, Kunkel, T.A. | | Deposit date: | 2008-01-31 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate-induced DNA strand misalignment during catalytic cycling by DNA polymerase lambda.
Embo Rep., 9, 2008
|
|
1HY3
 
 | | CRYSTAL STRUCTURE OF HUMAN ESTROGEN SULFOTRANSFERASE V269E MUTANT IN THE PRESENCE OF PAPS | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, ESTROGEN SULFOTRANSFERASE | | Authors: | Pedersen, L.C, Petrochenko, E.V, Shevtsov, S, Negishi, M. | | Deposit date: | 2001-01-17 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction.
J.Biol.Chem., 277, 2002
|
|
3C75
 
 | | Paracoccus versutus methylamine dehydrogenase in complex with amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, Methylamine dehydrogenase heavy chain, ... | | Authors: | Cavalieri, C, Biermann, N, Vlasie, M.D, Einsle, O, Merli, A, Ferrari, D, Rossi, G.L, Ubbink, M. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of crystal and solution states of the 138 kDa complex of methylamine dehydrogenase and amicyanin from Paracoccus versutus.
Biochemistry, 47, 2008
|
|
5OQU
 
 | | The crystal structure of CK2alpha in complex with compound 5 | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
1HXY
 
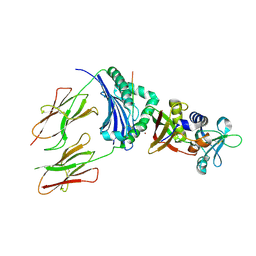 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H IN COMPLEX WITH HUMAN MHC CLASS II | | Descriptor: | ENTEROTOXIN H, HEMAGGLUTININ, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Petersson, K, Hakansson, M, Nilsson, H, Forsberg, G, Svensson, L.A, Liljas, A, Walse, B. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Superantigen Bound to MHC Class II Displays Zinc and Peptide Dependence
Embo J., 20, 2001
|
|
3C9P
 
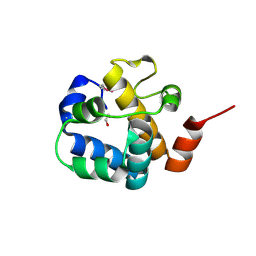 | | Crystal structure of uncharacterized protein SP1917 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein SP1917 | | Authors: | Chang, C, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of uncharacterized protein SP1917.
To be Published
|
|
1R0P
 
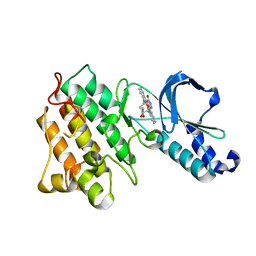 | | Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met in complex with the microbial alkaloid K-252a | | Descriptor: | Hepatocyte growth factor receptor, K-252A | | Authors: | Schiering, N, Knapp, S, Marconi, M, Flocco, M.M, Cui, J, Perego, R, Rusconi, L, Cristiani, C. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2Q7W
 
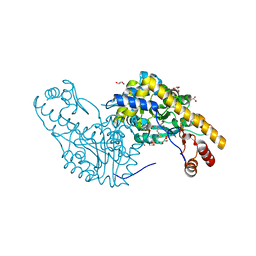 | | Structural Studies Reveals the Inactivation of E. coli L-aspartate aminotransferase (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms at pH 6.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
5OUE
 
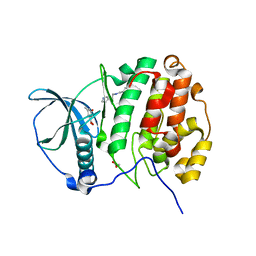 | | The crystal structure of CK2alpha in complex with compound 20 | | Descriptor: | (3-chloranyl-4-phenyl-phenyl)methyl-[2-(1~{H}-imidazol-4-yl)ethyl]azanium, 3-methyl-5-oxidanyl-benzoic acid, ACETATE ION, ... | | Authors: | Brear, P, De Fusco, C, Iegre, J, Yoshida, M, Mitchell, S, Rossmann, M, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Second-generation CK2 alpha inhibitors targeting the alpha D pocket.
Chem Sci, 9, 2018
|
|
6ZP7
 
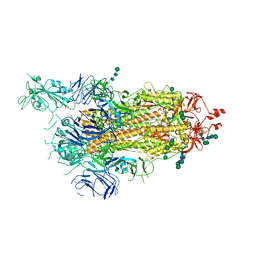 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
3BOM
 
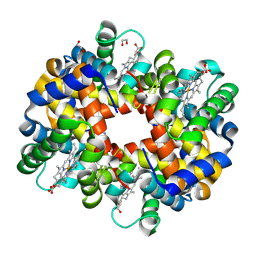 | | Crystal structure of trout hemoglobin at 1.35 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, Hemoglobin subunit alpha-4, Hemoglobin subunit beta-4, ... | | Authors: | Aranda IV, R, Bingman, C.A, Bitto, E, Wesenberg, G.E, Richards, M, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Trout Hemoglobin Crystal Structure at 1.35 Angstroms Resolution.
To be Published
|
|
1HJZ
 
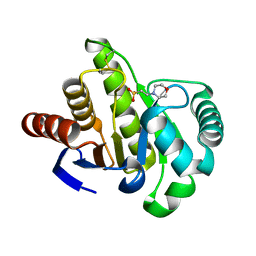 | | Crystal structure of AF1521 protein containing a macroH2A domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HYPOTHETICAL PROTEIN AF1521 | | Authors: | Allen, M.D, Buckle, A.M, Cordell, S.C, Lowe, J, Bycroft, M. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Af1521 a Protein from Archaeoglobus Fulgidus with Homology to the Non-Histone Domain of Macroh2A
J.Mol.Biol., 330, 2003
|
|
1HNH
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III + DEGRADED FORM OF ACETYL-COA | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, COENZYME A | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
1HND
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III-COA COMPLEX | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, COENZYME A | | Authors: | Qiu, X, Janson, C.A, Smith, W.W, Head, M, Lonsdale, J, Konstantinidis, A.K. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined structures of beta-ketoacyl-acyl carrier protein synthase III.
J.Mol.Biol., 307, 2001
|
|
6ZOW
 
 | | SARS-CoV-2 spike in prefusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
3ZQG
 
 | | Structure of Tetracycline repressor in complex with antiinducer peptide-TAP2 | | Descriptor: | ANTI-INDUCER PEPTIDE TAP2, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
6ZP5
 
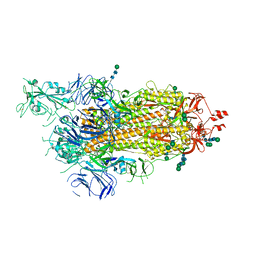 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
