6RIN
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
3KWB
 
 | | Structure of CatK covalently bound to a dioxo-triazine inhibitor | | Descriptor: | 3,5-dioxo-4-(3-piperidin-1-ylpropyl)-2-[3-(trifluoromethyl)phenyl]-2,3,4,5-tetrahydro-1,2,4-triazine-6-carbonitrile, Cathepsin K | | Authors: | Uitdehaag, J.C.M, van Zeeland, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Dioxo-triazines as a novel series of cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6KKV
 
 | |
6FMG
 
 | |
8HRG
 
 | | Tail tube of DT57C bacteriophage in the full state | | Descriptor: | Tail tube protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HOM
 
 | | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Omicron Main Protease (Mpro) in Complex with Ensitrelvir
To Be Published
|
|
5FL2
 
 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
8HQK
 
 | | Capsid of DT57C bacteriophage in the empty state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HRE
 
 | | Straight fiber of DT57C bacteriophage in the full state | | Descriptor: | Central straight fiber | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
5FG9
 
 | | Yeast 20S proteasome beta2-T(-2)V mutant | | Descriptor: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-12-20 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome.
Nat Commun, 7, 2016
|
|
8HOL
 
 | |
6RLY
 
 | | Human MMP12 (catalytic domain) in complex with AP316 | | Descriptor: | 2-(3-oxidanyl-2-oxidanylidene-pyridin-1-yl)-~{N}-[2-(4-phenylphenyl)ethyl]ethanamide, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Calderone, V, Fragai, M, Luchinat, C. | | Deposit date: | 2019-05-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploration of zinc-binding groups for the design of inhibitors for the oxytocinase subfamily of M1 aminopeptidases.
Bioorg.Med.Chem., 27, 2019
|
|
5FHS
 
 | |
5LNU
 
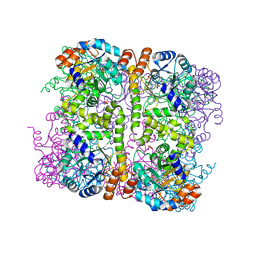 | | Crystal structure of Arabidopsis thaliana Pdx1-I320 complex | | Descriptor: | (4~{S})-4-azanyl-5-oxidanyl-pent-1-en-3-one, PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
2VEI
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE, SULFATE ION | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | Descriptor: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
5MLK
 
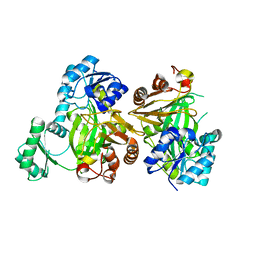 | |
2VEL
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties. | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, CHLORIDE ION, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
8HO3
 
 | | Capsid of DT57C bacteriophage in the full state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HQZ
 
 | | Baseplate of DT57C bacteriophage in the full state | | Descriptor: | Baseplate hub protein, Distal tail protein, L-shaped tail fiber assembly, ... | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
5MHL
 
 | |
2VKV
 
 | | TetR (BD) variant L17G with reverse phenotype | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, MAGNESIUM ION, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, ... | | Authors: | Resch, M, Striegl, H, Henssler, E.M, Sevvana, M, Egerer-Sieber, C, Schiltz, E, Hillen, W, Muller, Y.A. | | Deposit date: | 2008-01-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Protein Functional Leap: How a Single Mutation Reverses the Function of the Transcription Regulator Tetr.
Nucleic Acids Res., 36, 2008
|
|
8HOZ
 
 | |
1UYJ
 
 | | Clostridium perfringens epsilon toxin shows structural similarity with the pore forming toxin aerolysin | | Descriptor: | EPSILON-TOXIN, URANIUM ATOM | | Authors: | Cole, A.R, Gibert, M, Poppoff, M, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2004-03-02 | | Release date: | 2004-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Clostridium Perfringens Epsilon-Toxin Shows Structural Similarity to the Pore-Forming Toxin Aerolysin
Nat.Struct.Mol.Biol., 11, 2004
|
|
1G5F
 
 | | STRUCTURE OF LINB COMPLEXED WITH 1,2-DICHLOROETHANE | | Descriptor: | 1,2-DICHLOROETHANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, CALCIUM ION, ... | | Authors: | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
