5RC7
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library F11a | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCN
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 08 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RD3
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 25 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDK
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 44 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RE0
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 57 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7SUM
 
 | | Crystal structure of human ligase I with nick duplexes containing cognate A:T | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA ligase 1, DNA(5'-*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*A-3'), ... | | Authors: | Tang, Q, Gulkis, M, McKenna, R, Caglayan, M. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of LIG1 that engage with mutagenic mismatches inserted by pol beta in base excision repair.
Nat Commun, 13, 2022
|
|
7TRV
 
 | | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Crawford, M, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the DNA-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
6QJ3
 
 | | Crystal structure of the C. thermophilum condensin Ycs4-Brn1 subcomplex | | Descriptor: | Brn1, Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2 | | Authors: | Hassler, M, Haering, C.H, Kschonsak, M. | | Deposit date: | 2019-01-22 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of an Asymmetric Condensin ATPase Cycle.
Mol.Cell, 74, 2019
|
|
6C06
 
 | | Mycobacterium tuberculosis RNAP Holo/RbpA/Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
7TRW
 
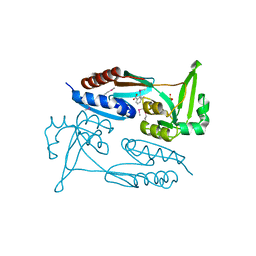 | | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis | | Descriptor: | 3-HYDROXYBENZOIC ACID, LysR-family transcriptional regulatory protein, PHOSPHATE ION | | Authors: | Kim, Y, Tesar, C, Crawford, M, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-31 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the C-terminal Ligand-Binding Domain of the LysR family Transcriptional Regulator YfbA from Yersinia pestis
To Be Published
|
|
7OA1
 
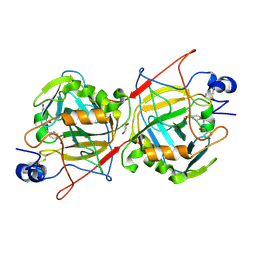 | |
6QJA
 
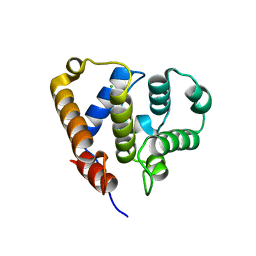 | | Organizational principles of the NuMA-Dynein interaction interface and implications for mitotic spindle functions | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nuclear mitotic apparatus protein 1 | | Authors: | Renna, C, Rizzelli, F, Carminati, M, Gaddoni, C, Pirovano, L, Cecatiello, V, Pasqualato, S, Mapelli, M. | | Deposit date: | 2019-01-23 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Organizational Principles of the NuMA-Dynein Interaction Interface and Implications for Mitotic Spindle Functions.
Structure, 28, 2020
|
|
4XOL
 
 | | Observing the overall rocking motion of a protein in a crystal - Cubic Ubiquitin crystals. | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Coquelle, N, Peixiang, M, Schanda, P, Colletier, J.P. | | Deposit date: | 2015-01-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Observing the overall rocking motion of a protein in a crystal.
Nat Commun, 6, 2015
|
|
7TKV
 
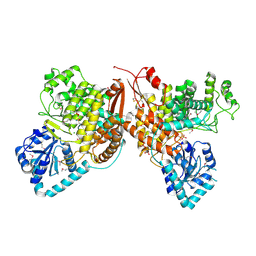 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-17 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
6W85
 
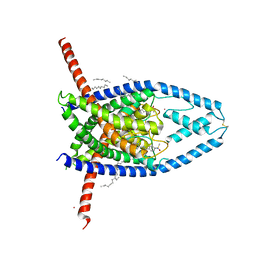 | | K2P2.1 (TREK-1):ML335 complex, 200 mM K+ | | Descriptor: | CADMIUM ION, DECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
5WR8
 
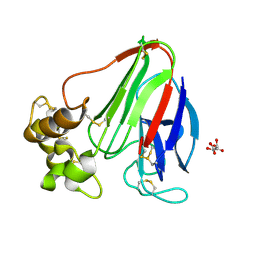 | | Thaumatin structure determined by SACLA at 1.55 Angstrom | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Sugahara, M. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
6W8C
 
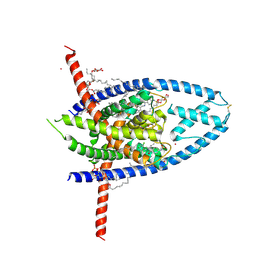 | | K2P2.1 (TREK-1):ML335 complex, 1 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
6QF5
 
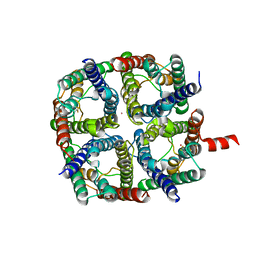 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QGO
 
 | | Crystal structure of APT1 S119A mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
5LG3
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.567 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7JX4
 
 | | Crystal Structure of N-Lysine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Lysine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices.
J.Am.Chem.Soc., 143, 2021
|
|
7JX5
 
 | | Crystal Structure of N-Phenylalanine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Phenylalanine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices
J.Am.Chem.Soc., 143, 2021
|
|
4XQ3
 
 | | Crystal structure of Sso-SmAP2 | | Descriptor: | Like-Sm ribonucleoprotein core | | Authors: | Bezerra, G.A, Martens, B, Kreuter, M.J, Grishkovskaya, I, Manica, M, Arkhipova, V, Blasi, U, Djinovic-Carugo, K. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets.
Life, 5, 2015
|
|
6O9R
 
 | |
2G4I
 
 | | Anomalous substructure of Concanavalin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Concanavalin A, ... | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
