5OVK
 
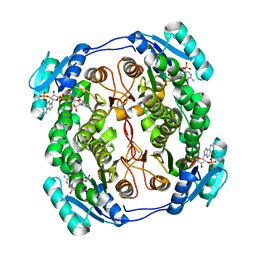 | | Crystal structure MabA bound to NADPH from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3NNH
 
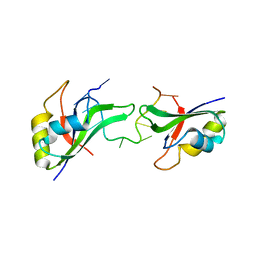 | | Crystal Structure of the CUGBP1 RRM1 with GUUGUUUUGUUU RNA | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7501 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
6HL0
 
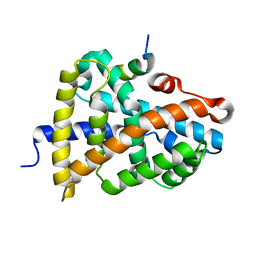 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide | | Descriptor: | Bile acid receptor, NCoA-2 peptide (Nuclear receptor coactivator 2), LYS-GLU-ASN-ALA-LEU-LEU-ARG-TYR-LEU-LEU-ASP-LYS-ASP | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
6HTX
 
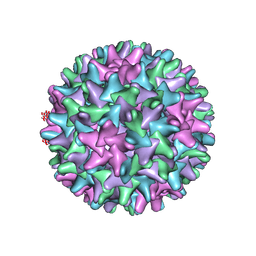 | | WT Hepatitis B core protein capsid | | Descriptor: | Capsid protein | | Authors: | Bottcher, B, Nassal, M. | | Deposit date: | 2018-10-05 | | Release date: | 2018-12-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of Mutant Hepatitis B Core Protein Capsids with Premature Secretion Phenotype.
J. Mol. Biol., 430, 2018
|
|
6HLA
 
 | | wild-type NuoEF from Aquifex aeolicus - reduced form bound to NADH | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HMG
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
3NOO
 
 | | Crystal Structure of C101A Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ThiJ/PfpI family protein | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
6HN7
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC), Terminase small subunit | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
6HOL
 
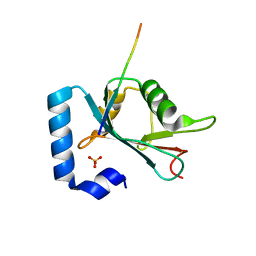 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | Descriptor: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
3NUR
 
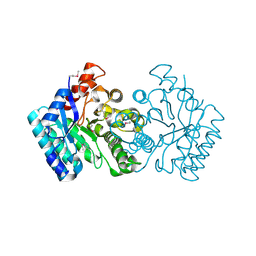 | | Crystal structure of a putative amidohydrolase from Staphylococcus aureus | | Descriptor: | Amidohydrolase, CALCIUM ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Lam, K, Soloveychik, M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative amidohydrolase from Staphylococcus aureus
To be Published
|
|
3NV5
 
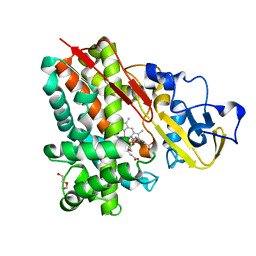 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
6HT4
 
 | |
8CK3
 
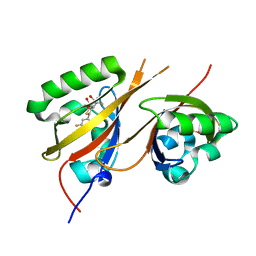 | | STRUCTURE OF HIF2A-ARNT HETERODIMER IN COMPLEX WITH (S)-1-(3,5-Difluoro-phenyl)-5,5-difluoro-3-methanesulfonyl-5,6-dihydro-4H-cyclopenta[c]thiophen-4-ol | | Descriptor: | (4~{S})-1-[3,5-bis(fluoranyl)phenyl]-5,5-bis(fluoranyl)-3-methylsulfonyl-4,6-dihydrocyclopenta[c]thiophen-4-ol, Aryl hydrocarbon receptor nuclear translocator, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Lehmannn, M, Diehl, L. | | Deposit date: | 2023-02-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Discovery of Cycloalkyl[ c ]thiophenes as Novel Scaffolds for Hypoxia-Inducible Factor-2 alpha Inhibitors.
J.Med.Chem., 66, 2023
|
|
6HRI
 
 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
3NW4
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans in complex with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
5OXU
 
 | |
6HYH
 
 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Fucofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, ZINC ION, beta-D-fucofuranose | | Authors: | Li, M, Mueller, C, Zhang, L, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-10-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HYW
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT Y119F | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3NVC
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans in complex with salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
3O75
 
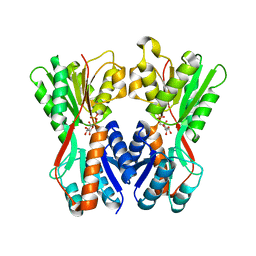 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
6I0S
 
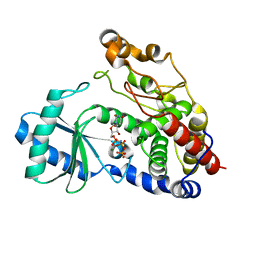 | | Crystal structure of DmTailor in complex with UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
6HZL
 
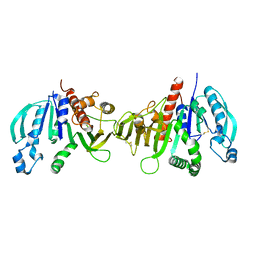 | | Crystal structure of redox-inhibited phosphoribulokinase from Synechococcus sp. (strain PCC 6301), osmate derivative | | Descriptor: | OSMIUM ION, Phosphoribulokinase | | Authors: | Wilson, R.H, Bracher, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6I0T
 
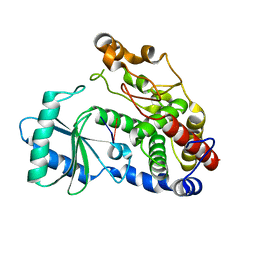 | | Crystal structure of DmTailor in complex with GpU | | Descriptor: | RNA (5'-R(*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
3O4K
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) and lipoteichoic acid at 2.1 A resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of recognition of pathogen-associated molecular patterns and inhibition of proinflammatory cytokines by camel peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
6HCB
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.9 A RESOLUTION. | | Descriptor: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Masternak, M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
