6XQ9
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 13 | | Descriptor: | 4-(3-hydroxyphenoxy)benzoic acid, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ1
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-[3-({[3-(4-carboxyphenoxy)phenyl]methoxy}methyl)phenyl]-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
2DVQ
 
 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | Bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
6XU6
 
 | | Drosophila melanogaster Testis 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6XU8
 
 | | Drosophila melanogaster Ovary 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6XU7
 
 | | Drosophila melanogaster Testis polysome ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6X6N
 
 | |
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y0H
 
 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y94
 
 | | Ca2+-bound Calmodulin mutant N53I | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Holt, C, Nielsen, L.H, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6Y3Z
 
 | |
6Y1L
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47R mutant | | Descriptor: | DIETHYL PHOSPHONATE, FAB A.17 L47R mutant HEAVY CHAIN, FAB A.17 L47R mutant Light CHAIN, ... | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y4P
 
 | | Calmodulin N53I variant bound to cardiac ryanodine receptor (RyR2) calmodulin binding domain | | Descriptor: | CALCIUM ION, Calmodulin-1, Ryanodine receptor 2 | | Authors: | Lau, K, Nielsen, L.H, Holt, C, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Van Petegem, F, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13325572 Å) | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6Y3P
 
 | |
6Y6K
 
 | | Cryo-EM structure of a Phenuiviridae L protein | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase | | Authors: | Vogel, D, Thorkelsson, S.R, Quemin, E, Meier, K, Kouba, T, Gogrefe, N, Busch, C, Reindl, S, Guenther, S, Cusack, S, Gruenewald, K, Rosenthal, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
6Y9M
 
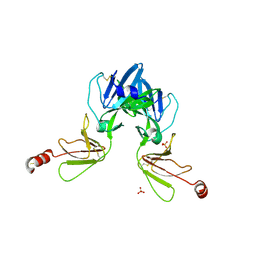 | | Crystal structure of TSWV glycoprotein N ectodomain (sGn) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, PHOSPHATE ION, ... | | Authors: | Dessau, M, Bahat, Y. | | Deposit date: | 2020-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of tomato spotted wilt virus G N reveals a dimer complex formation and evolutionary link to animal-infecting viruses
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y49
 
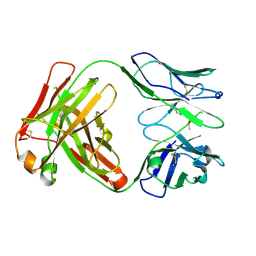 | | Crystal structure of the paraoxon-modified A.17kappa antibody FAB fragment | | Descriptor: | A.17kappa antibody FAB fragment - Heavy Chain, A.17kappa antibody FAB fragment - Light Chain, DIETHYL PHOSPHONATE | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-19 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y33
 
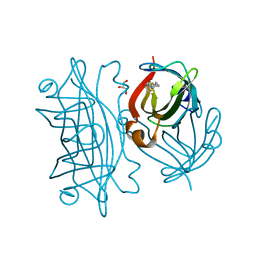 | | Streptavidin mutant S112R with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6YB9
 
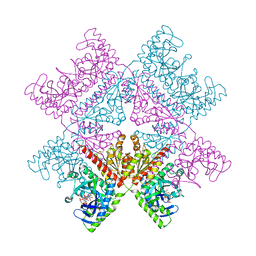 | | Human octameric PAICS in complex with SAICAR, AMP-PNP, and magnesium | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Skerlova, J, Unterlass, J, Gottmann, M, Homan, E, Helleday, T, Jemth, A.S, Stenmark, P. | | Deposit date: | 2020-03-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target.
J.Biol.Chem., 295, 2020
|
|
6YD0
 
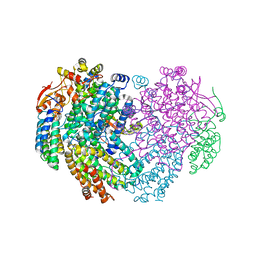 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, diferric state | | Descriptor: | FE (III) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
3CI1
 
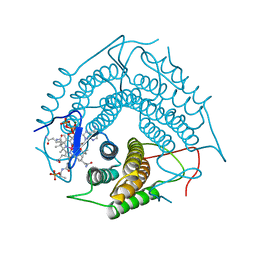 | | Structure of the PduO-type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri complexed with four-coordinate cob(II)alamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | Maurice, M.St, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a human-type corrinoid adenosyltransferase confirms that coenzyme B12 is synthesized through a four-coordinate intermediate.
Biochemistry, 47, 2008
|
|
6XZM
 
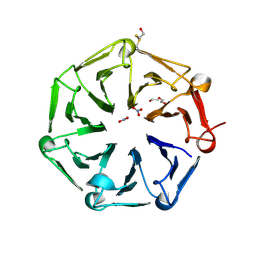 | | Arabidopsis UV-B photoreceptor UVR8 mutant D96N D107N W285A | | Descriptor: | DI(HYDROXYETHYL)ETHER, NITRATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Lau, K, Hothorn, M. | | Deposit date: | 2020-02-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.10009313 Å) | | Cite: | A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Y1N
 
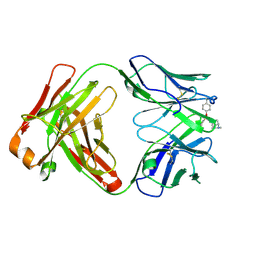 | | Crystal structure of the phosphonate-modified A.5 antibody FAB fragment | | Descriptor: | 8-METHYL-8-AZABICYCLO[3.2.1]OCTAN-3-YL PHENYLPHOSPHONATE, FAB A.5 Heavy chain, FAB A.5 Light Chain | | Authors: | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2020-02-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Y2O
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with 1,7-Naphthyridin-8-amine and PKI (5-24) | | Descriptor: | 1,7-naphthyridin-8-amine, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Oebbeke, M, Heine, A, Klebe, G. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
