7K19
 
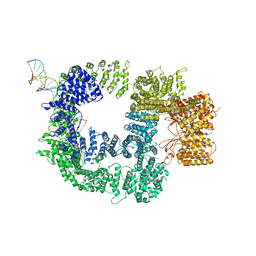 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex I) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
1UX0
 
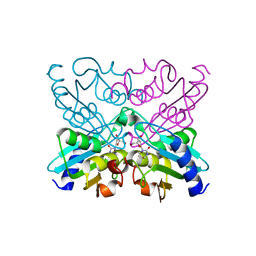 | | Bacillus subtilis cytidine deaminase with an Arg56 - Gln substitution | | Descriptor: | CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ZINC ION | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1UPR
 
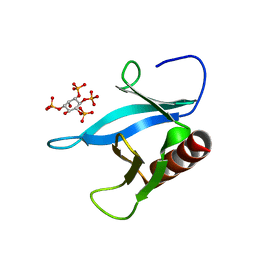 | | Crystal structure of the PEPP1 pleckstrin homology domain in complex with Inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, PLECKSTRIN HOMOLOGY DOMAIN-CONTAINING FAMILY A MEMBER 4 | | Authors: | Milburn, C.C, Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Pleckstrin Homology Domain of Pepp1
To be Published
|
|
7JLX
 
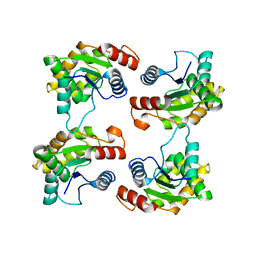 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ (TIR domains) | | Descriptor: | Disease resistance protein Roq1 | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7K37
 
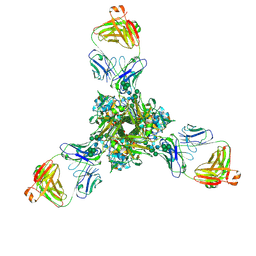 | | Structure of full-length influenza HA with a head-binding antibody at pH 7.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Xiang, Y, Gao, J. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
4X28
 
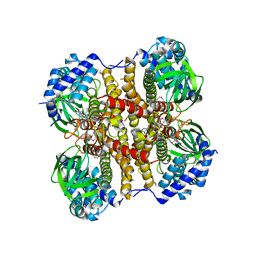 | | Crystal structure of the ChsE4-ChsE5 complex from Mycobacterium tuberculosis | | Descriptor: | Acyl-CoA dehydrogenase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Unraveling Cholesterol Catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 alpha 2 beta 2 Acyl-CoA Dehydrogenase Initiates beta-Oxidation of 3-Oxo-cholest-4-en-26-oyl CoA.
Acs Infect Dis., 1, 2015
|
|
1URH
 
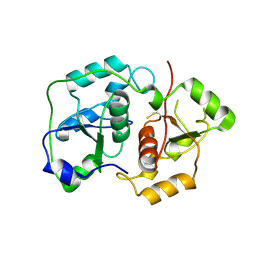 | | The "Rhodanese" fold and catalytic mechanism of 3-mercaptopyruvate sulfotransferases: Crystal structure of SseA from Escherichia coli | | Descriptor: | 3-MERCAPTOPYRUVATE SULFURTRANSFERASE, SULFITE ION | | Authors: | Spallarossa, A, Forlani, F, Carpen, A, Armirotti, A, Pagani, S, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-10-30 | | Release date: | 2003-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The "Rhodanese" Fold and Catalytic Mechanism of 3-Mercaptopyruvate Sulfurtransferases: Crystal Structure of Ssea from Escherichia Coli
J.Mol.Biol., 335, 2004
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
1UX1
 
 | | Bacillus subtilis cytidine deaminase with a Cys53His and an Arg56Gln substitution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ... | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
4CBT
 
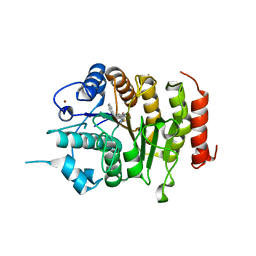 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | Descriptor: | (1R,2R,3R)-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, ZINC ION | | Authors: | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
7K0T
 
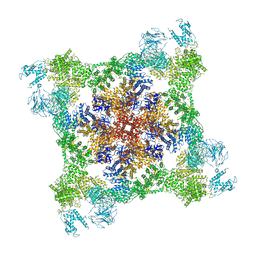 | |
1V02
 
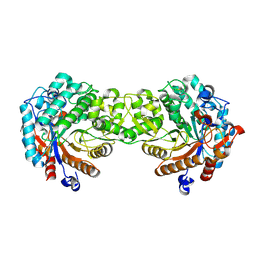 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | DHURRINASE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
7K0S
 
 | |
1UYW
 
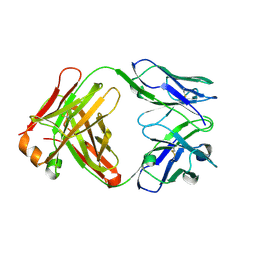 | | Crystal Structure of the antiflavivirus Fab4g2 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Taylor, E.J, Gil-Valdes, J, Chinea, G, Davies, G. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Antiflavivirus Fab4G2
To be Published
|
|
5NUK
 
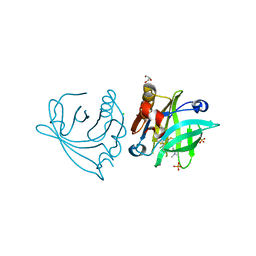 | | Engineered beta-lactoglobulin: variant I56F-L39A-M107F in complex with chlorpromazine (LG-FAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin, CHLORIDE ION, ... | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Lazinska, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
1VBS
 
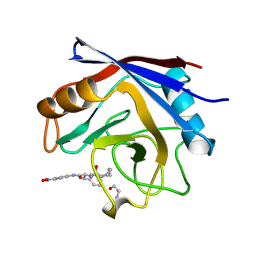 | | STRUCTURE OF CYCLOPHILIN COMPLEXED WITH (D)ALA CONTAINING TETRAPEPTIDE | | Descriptor: | CYCLOPHILIN A, TETRAPEPTIDE | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the stereospecificity of peptidyl prolyl cis/trans isomerases.
FEBS Lett., 432, 1998
|
|
1VBT
 
 | | Structure of cyclophilin complexed with sulfur-substituted tetrapeptide AAPF | | Descriptor: | CYCLOPHILIN A, SULFUR-SUBSTITUTED TETRAPEPTIDE | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into Conversion of Substrate to Inhibitor
To be Published
|
|
1UT0
 
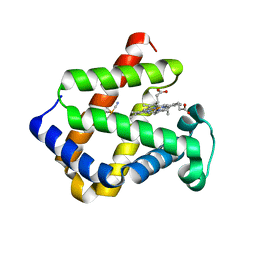 | | CRYSTAL STRUCTURE OF CYTOGLOBIN: THE FOURTH GLOBIN TYPE DISCOVERED IN MAN DISPLAYS HEME HEXA-COORDINATION | | Descriptor: | CYTOGLOBIN, HEXACYANOFERRATE(3-), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | De Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Cytoglobin: The Fourth Globin Type Discovered in Man Displays Heme Hexa-Coordination
J.Mol.Biol., 336, 2004
|
|
7JRK
 
 | |
5O1U
 
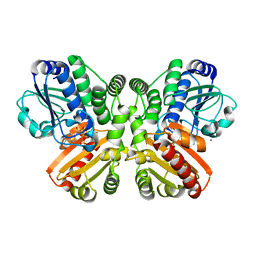 | | Structure of wildtype T.maritima PDE (TM1595) with AMP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-19 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
1URJ
 
 | | Single stranded DNA-binding protein(ICP8) from Herpes simplex virus-1 | | Descriptor: | MAJOR DNA-BINDING PROTEIN, MERCURY (II) ION, ZINC ION | | Authors: | Panjikar, S, Mapelli, M, Tucker, P.A. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding.
J. Biol. Chem., 280, 2005
|
|
7K1U
 
 | | Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Collagen-binding Adhesin | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of SrtB-anchored Collagen-binding Adhesin Fragment (residues 206-565) from Clostridioides difficile strain 630
To Be Published
|
|
1UW7
 
 | | Nsp9 protein from SARS-coronavirus. | | Descriptor: | NSP9 | | Authors: | Sutton, G, Fry, E, Carter, L, Sainsbury, S, Walter, T, Nettleship, J, Berrow, N, Owens, R, Gilbert, R, Davidson, A, Siddell, S, Poon, L.L.M, Diprose, J, Alderton, D, Walsh, M, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Nsp9 Replicase Protein of Sars-Coronavirus, Structure and Functional Insights
Structure, 12, 2004
|
|
1V0P
 
 | | Structure of P. falciparum PfPK5-Purvalanol B ligand complex | | Descriptor: | CELL DIVISION CONTROL PROTEIN 2 HOMOLOG, PURVALANOL B | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
1OSB
 
 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | Dna oligonucleotide, SULFATE ION, TrwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
