3NQ4
 
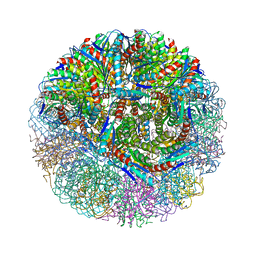 | |
4HVA
 
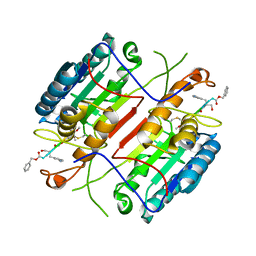 | |
2XTZ
 
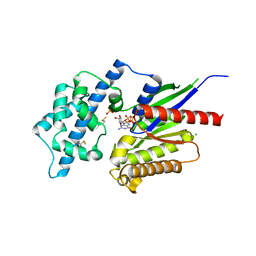 | | Crystal structure of the G alpha protein AtGPA1 from Arabidopsis thaliana | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, GUANINE NUCLEOTIDE-BINDING PROTEIN ALPHA-1 SUBUNIT, ... | | Authors: | Jones, J.C, Duffy, J.W, Machius, M, Temple, B.R.S, Dohlman, H.G, Jones, A.M. | | Deposit date: | 2010-10-13 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The Crystal Structure of a Self-Activating G Protein Alpha Subunit Reveals its Distinct Mechanism of Signal Initiation
Sci.Signal., 159, 2011
|
|
2A5L
 
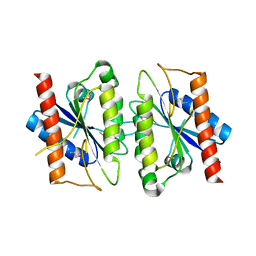 | | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Trp repressor binding protein WrbA | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa
To be Published
|
|
2XYT
 
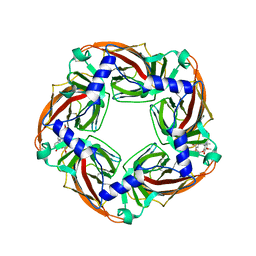 | | Crystal structure of Aplysia californica AChBP in complex with d- tubocurarine | | Descriptor: | D-TUBOCURARINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
4HW8
 
 | | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose. | | Descriptor: | Bacterial extracellular solute-binding protein, putative, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose.
TO BE PUBLISHED
|
|
2Y1P
 
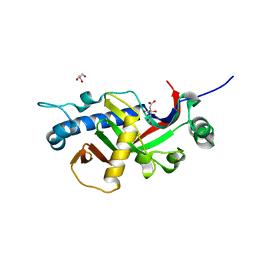 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, complexed with citrate | | Descriptor: | 2', 3'-CYCLIC NUCLEOTIDE 3'-PHOSPHODIESTERASE, CITRIC ACID, ... | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2010-12-09 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Myelin 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase: Active-Site Ligand Binding and Molecular Conformation.
Plos One, 7, 2012
|
|
2A9K
 
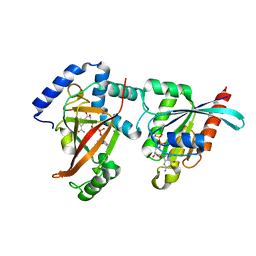 | | Crystal structure of the C3bot-NAD-RalA complex reveals a novel type of action of a bacterial exoenzyme | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mono-ADP-ribosyltransferase C3, ... | | Authors: | Pautsch, A, Vogelsgesang, M, Trankle, J, Herrmann, C, Aktories, K. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme.
Embo J., 24, 2005
|
|
4HY6
 
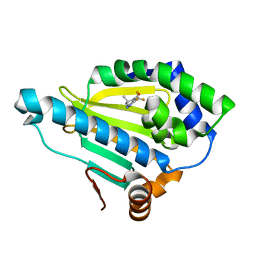 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1 | | Descriptor: | 6,6-dimethyl-3-(trifluoromethyl)-1,5,6,7-tetrahydro-4H-indazol-4-one, Heat shock protein HSP 90-alpha | | Authors: | Yang, M, Li, J, He, J.H. | | Deposit date: | 2012-11-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ1
to be published
|
|
4HZ4
 
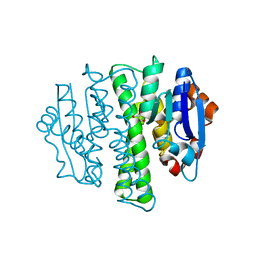 | | Crystal structure of glutathione s-transferase b4xh91 (target efi-501787) from actinobacillus pleuropneumoniae | | Descriptor: | GLYCEROL, Glutathione-S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of glutathione s-transferase b4xh91 from Actinobacillus pleuropneumoniae
To be Published
|
|
3OGK
 
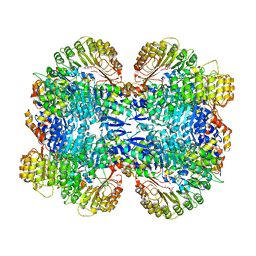 | | Structure of COI1-ASK1 in complex with coronatine and an incomplete JAZ1 degron | | Descriptor: | (1S,2S)-2-ethyl-1-({[(3aS,4S,6R,7aS)-6-ethyl-1-oxooctahydro-1H-inden-4-yl]carbonyl}amino)cyclopropanecarboxylic acid, Coronatine-insensitive protein 1, JAZ1 incomplete degron peptide, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
2XQ2
 
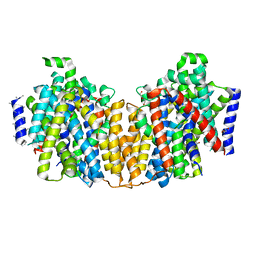 | | Structure of the K294A mutant of vSGLT | | Descriptor: | DI(HYDROXYETHYL)ETHER, SODIUM/GLUCOSE COTRANSPORTER | | Authors: | Watanabe, A, Choe, S, Chaptal, V, Rosenberg, J.M, Wright, E.M, Grabe, M, Abramson, J. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Mechanism of Sodium and Substrate Release from the Binding Pocket of Vsglt
Nature, 468, 2010
|
|
2XR1
 
 | | DIMERIC ARCHAEAL CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR WITH N-TERMINAL KH DOMAINS (KH-CPSF) FROM METHANOSARCINA MAZEI | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 100 KD SUBUNIT, ZINC ION | | Authors: | Mir-Montazeri, B, Ammelburg, M, Forouzan, D, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2010-09-08 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Dimeric Archaeal Cleavage and Polyadenylation Specificity Factor.
J.Struct.Biol., 173, 2011
|
|
2ADD
 
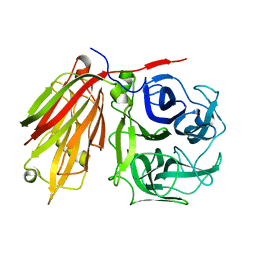 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
3OC3
 
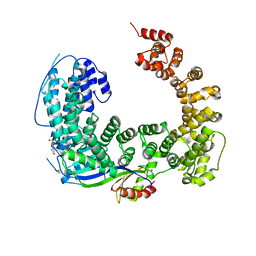 | | Crystal structure of the Mot1 N-terminal domain in complex with TBP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HELICASE MOT1, TRANSCRIPTION INITIATION FACTOR TFIID (TFIID-1) | | Authors: | Wollmann, P, Cui, S, Viswanathan, R, Berninghausen, O, Wells, M.N, Moldt, M, Witte, G, Butryn, A, Wendler, P, Beckmann, R, Auble, D.T, Hopfner, K.-P. | | Deposit date: | 2010-08-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP.
Nature, 475, 2011
|
|
2Y5L
 
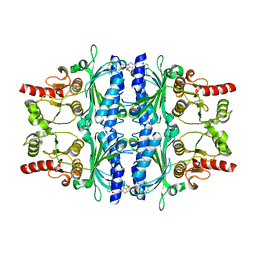 | | orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-bromo-1,3-thiazol-2(3H)-ylidene]carbamoyl}-3-chlorobenzenesulfonamide | | Authors: | ruf, a, hebeisen, p, haap, w, kuhn, b, mohr, p, wessel, h.p, zutter, u, kirchner, s, benz, j, joseph, c, alvarez-sanchez, r, gubler, m, schott, b, benardeau, a, tozzo, e, kitas, e. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2AFG
 
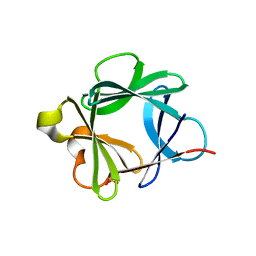 | |
1ZXT
 
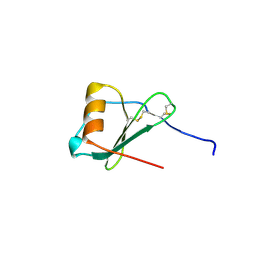 | | Crystal Structure of A Viral Chemokine | | Descriptor: | functional macrophage inflammatory protein 1-alpha homolog | | Authors: | Luz, J.G, Yu, M, Su, Y, Wu, Z, Zhou, Z, Sun, R, Wilson, I.A. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of viral macrophage inflammatory protein I encoded by Kaposi's sarcoma-associated herpesvirus at 1.7A.
J.Mol.Biol., 352, 2005
|
|
1ZZI
 
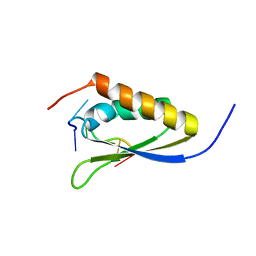 | | Crystal Structure Analysis of the third KH domain of hnRNP K in complex with ssDNA | | Descriptor: | 5'-D(*CP*TP*CP*CP*CP*C)-3', Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
2A5J
 
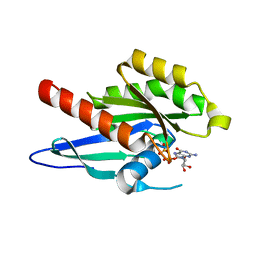 | | Crystal Structure of Human RAB2B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-2B | | Authors: | Dong, A, Wang, J, Shen, Y, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of human RAB2B
To be Published
|
|
4HZ1
 
 | | Crystal Structure of Pseudomonas aeruginosa azurin with iron(II) at the copper-binding site. | | Descriptor: | ACETATE ION, Azurin, FE (II) ION | | Authors: | McLaughlin, M.P, Retegan, M, Bill, E, Payne, T.M, Shafaat, H.S, Pea, S, Sudhamsu, J, Ensign, A.A, Crane, B.R, Neese, F, Holland, P.L. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azurin as a Protein Scaffold for a Low-coordinate Nonheme Iron Site with a Small-molecule Binding Pocket.
J.Am.Chem.Soc., 134, 2012
|
|
2A5U
 
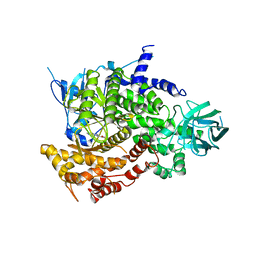 | | Crystal Structure of human PI3Kgamma complexed with AS605240 | | Descriptor: | (5E)-5-(QUINOXALIN-6-YLMETHYLENE)-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
3OD9
 
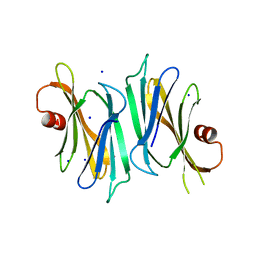 | | Crystal structure of PliI-Ah, periplasmic lysozyme inhibitor of I-type lysozyme from Aeromonas hydrophyla | | Descriptor: | POTASSIUM ION, Putative exported protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-11 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
2AHI
 
 | | Structural Basis of DNA Recognition by p53 Tetramers (complex III) | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*AP*TP*GP*TP*CP*CP*G)-3', Cellular tumor antigen p53, ZINC ION | | Authors: | Kitayner, M, Rozenberg, H, Kessler, N, Rabinovich, D, Shakked, Z. | | Deposit date: | 2005-07-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of DNA Recognition by p53 Tetramers
Mol.Cell, 22, 2006
|
|
3OGX
 
 | | Crystal structure of the complex of Peptidoglycan Recognition protein (PGRP-s) with Heparin-Dissacharide at 2.8 A resolution | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of heparin binding to camel peptidoglycan recognition protein-S
Int J Biochem Mol Biol, 3, 2012
|
|
