1ZEE
 
 | | X-Ray Crystal Structure of Protein SO4414 from Shewanella oneidensis. Northeast Structural Genomics Consortium Target SoR52. | | Descriptor: | hypothetical protein SO4414 | | Authors: | Forouhar, F, Abashidze, M, Vorobiev, S.M, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of the Hypothetical Protein SO4414 from Shewanella oneidensis, NESG Target SoR52
To be Published
|
|
2WY4
 
 | | Structure of bacterial globin from Campylobacter jejuni at 1.35 A resolution | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SINGLE DOMAIN HAEMOGLOBIN | | Authors: | Barynin, V.V, Sedelnikova, S.E, Shepherd, M, Wu, G, Poole, R.K, Rice, D.W. | | Deposit date: | 2009-11-11 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Single-Domain Globin from the Pathogenic Bacterium Campylobacter Jejuni: Novel D-Helix Conformation, Proximal Hydrogen Bonding that Influences Ligand Binding, and Peroxidase-Like Redox Properties.
J.Biol.Chem., 285, 2010
|
|
2WZD
 
 | | The catalytically active fully closed conformation of human phosphoglycerate kinase K219A mutant in complex with ADP, 3PG and aluminium trifluoride | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Bowler, M.W, Cliff, M.J, Marston, J.P.M, Baxter, N.J, Hownslow, A.M.H, Varga, A.V, Szabo, J, Vas, M, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Transition State Analogue Structures of Human Phosphoglycerate Kinase Establish the Importance of Charge Balance in Catalysis.
J.Am.Chem.Soc., 132, 2010
|
|
3P5T
 
 | | CFIm25-CFIm68 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
2WPU
 
 | | Chaperoned ruthenium metallodrugs that recognize telomeric DNA | | Descriptor: | (3AS,4S,6AR)-4-(5-((3R,4R)-3,4-DIAMINOPYRROLIDIN-1-YL)-5-OXOPENTYL)TETRAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-2(3H)-ONE-P-CYMENE-CHLORO-RUTHENIUM(III), GLYCEROL, STREPTAVIDIN, ... | | Authors: | Heinisch, T, Schirmer, T, Zimbron, J.M, Sardo, A, Wohlschlager, T, Gradinaru, J, Creus, M, Ward, T.R. | | Deposit date: | 2009-08-10 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Chemo-Genetic Optimization of DNA Recognition by Metallodrugs Using a Presenter-Protein Strategy.
Chemistry, 16, 2010
|
|
8BZ1
 
 | | RNA polymerase II core pre-initiation complex with the proximal +1 nucleosome (cPIC-Nuc10W) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
3P7V
 
 | |
5B4U
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor malonate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, CHLORIDE ION, MALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the catalytic reaction trigger and inhibition of D-3-hydroxybutyrate dehydrogenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2WZQ
 
 | | Insertion Mutant E173GP174 of the NS3 protease-helicase from dengue virus | | Descriptor: | CHLORIDE ION, GLYCEROL, NS3 PROTEASE-HELICASE | | Authors: | Luo, D, Wei, N, Doan, D, Paradkar, P, Chong, Y, Davidson, A, Kotaka, M, Lescar, J, Vasudevan, S. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility between the Protease and Helicase Domains of the Dengue Virus Ns3 Protein Conferred by the Linker Region and its Functional Implications.
J.Biol.Chem., 285, 2010
|
|
3PA9
 
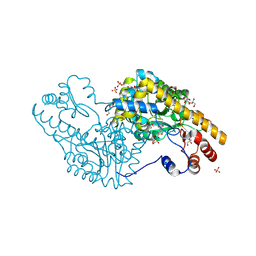 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 7.5 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
5B5L
 
 | | Crystal structure of acetyl esterase mutant S10A with acetate ion | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uechi, K, Kamachi, S, Akita, H, Mine, S, Watanabe, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | crystal structure of acetyl esterase mutant S10A with acetate ion
To Be Published
|
|
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
4GXA
 
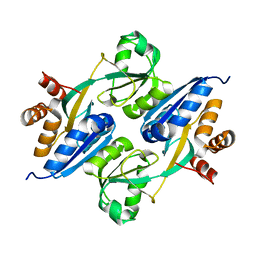 | |
3OWH
 
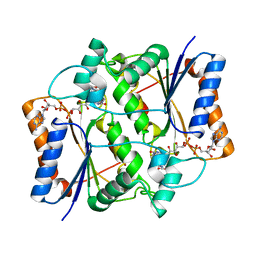 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Mesecar, A.D. | | Deposit date: | 2010-09-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
5B6O
 
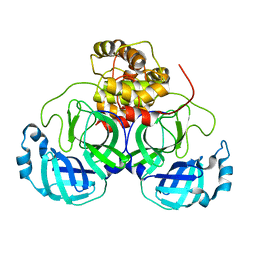 | | Crystal structure of MS8104 | | Descriptor: | 3C-like proteinase | | Authors: | Wang, H, Kim, Y, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4GYD
 
 | | Nostoc sp Cytochrome c6 | | Descriptor: | Cytochrome c6, HEME C | | Authors: | Skubak, P, Ubbink, M, Cavazzini, D, Rossi, G.L, Pannu, N.S. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy
Biochim.Biophys.Acta, 1837, 2014
|
|
3ONW
 
 | | Structure of a G-alpha-i1 mutant with enhanced affinity for the RGS14 GoLoco motif. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, Regulator of G-protein signaling 14, ... | | Authors: | Bosch, D, Kimple, A.J, Sammond, D.W, Miley, M.J, Machius, M, Kuhlman, B, Willard, F.S, Siderovski, D.P. | | Deposit date: | 2010-08-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Determinants of Affinity Enhancement between GoLoco Motifs and G-Protein {alpha} Subunit Mutants.
J.Biol.Chem., 286, 2011
|
|
5BQ9
 
 | | Crystal structure of uncharacterized protein lpg1496 Legionella pneumophila subsp. pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Morar, M, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.2785 Å) | | Cite: | Crystal structure of the Legionella pneumophila lem10 effector reveals a new member of the HD protein superfamily.
Proteins, 83, 2015
|
|
3OOI
 
 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
4GXZ
 
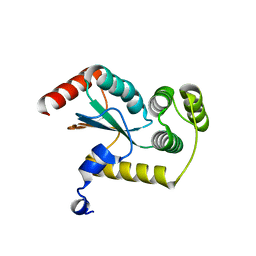 | | Crystal structure of a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium | | Descriptor: | Suppression of copper sensitivity protein | | Authors: | Shepherd, M, Heras, B, King, G.J, Argente, M.P, Achard, M.E.S, King, N.P, McEwan, A.G, Schembri, M.A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium
Antioxid Redox Signal, 19, 2013
|
|
4GYR
 
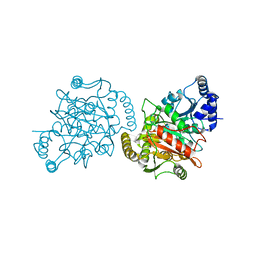 | | Granulibacter bethesdensis allophanate hydrolase apo | | Descriptor: | Allophanate hydrolase | | Authors: | Lin, Y, St Maurice, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Allophanate Hydrolase from Granulibacter bethesdensis Provides Insights into Substrate Specificity in the Amidase Signature Family.
Biochemistry, 52, 2013
|
|
1XV6
 
 | |
4GYI
 
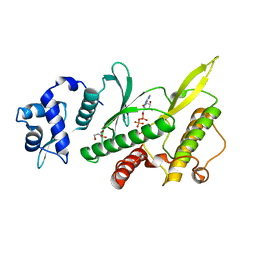 | | Crystal structure of the Rio2 kinase-ADP/Mg2+-phosphoaspartate complex from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ferreira-Cerca, S, Sagar, V, Schafer, T, Diop, M, Wesseling, A.M, Lu, H, Chai, E, Hurt, E, LaRonde-LeBlanc, N. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2X80
 
 | | P450 BM3 F87A in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450/NADPH-P450 REDUCTASE, DIMETHYL SULFOXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Wong, T.S, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2010-03-05 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Role of Active-Site Phe87 in Modulating the Organic Co-Solvent Tolerance of Cytochrome P450 Bm3 Monooxygenase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8BVW
 
 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
