3RMH
 
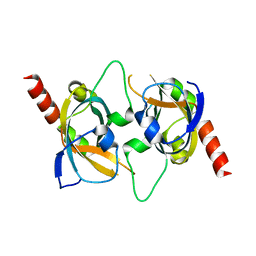 | | Crystal Structure of yeast telomere protein Cdc13 OB4 | | Descriptor: | Yeast Cdc13 OB4 | | Authors: | Sun, J, Yang, Y, Lei, M. | | Deposit date: | 2011-04-20 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Analyses of Candida Cdc13 Orthologues Revealed a Novel OB Fold Dimer Arrangement, Dimerization-Assisted DNA Binding, and Substantial Structural Differences between Cdc13 and RPA70.
Mol.Cell.Biol., 32, 2012
|
|
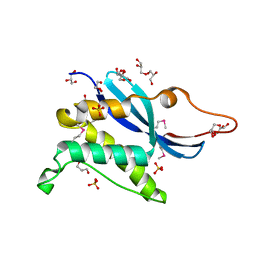
1KQ6
 
 | |
3R4X
 
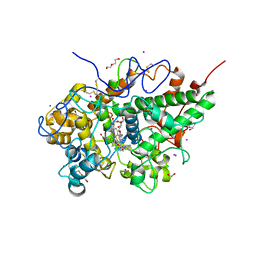 | | Crystal structure of bovine lactoperoxidase complexed with pyrazine-2-carboxamide at 2 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of bovine lactoperoxidase complexed with Pyrazine-2-carboxamide at 2 A resolution
To be Published
|
|
6JY6
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
4MKN
 
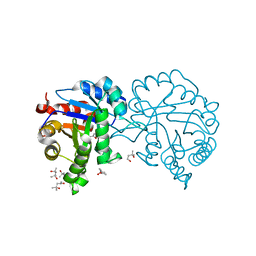 | | Crystal structure of chloroplastic triosephosphate isomerase from Chlamydomonas reinhardtii at 1.1 A of resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Triosephosphate isomerase | | Authors: | Fermani, S, Sciabolini, C, Zaffagnini, M, Lemaire, S.D. | | Deposit date: | 2013-09-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Crystal Structure and Redox Properties of Chloroplastic Triosephosphate Isomerase from Chlamydomonas reinhardtii.
Mol Plant, 7, 2014
|
|
6JYC
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
3R5W
 
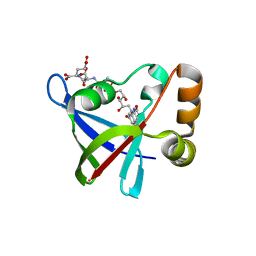 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
4LBF
 
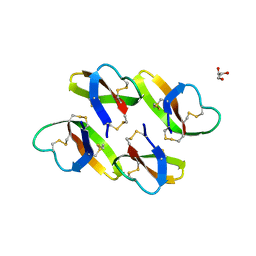 | |
6TTI
 
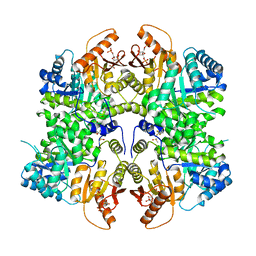 | | PKM2 in complex with Compound 6 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, DIMETHYL SULFOXIDE, Pyruvate kinase PKM, ... | | Authors: | Saur, M, Hartshorn, M.J, Dong, J, Reeks, J, Bunkoczi, G, Jhoti, H, Williams, P.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Fragment-based drug discovery using cryo-EM.
Drug Discov Today, 25, 2020
|
|
4LBQ
 
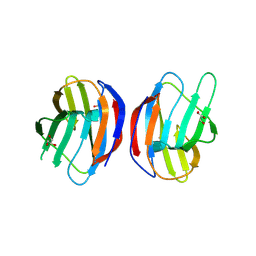 | | Crystal structure of mouse galectin-1 | | Descriptor: | GLYCEROL, Galectin-1 | | Authors: | Rustiguel, J.K, Trabuco, A.C, Del Cistia Andrade, C, Stowell, S.R, Cummings, R.D, Dias-Baruffi, M, Nonato, M.C. | | Deposit date: | 2013-06-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of mouse galectin-1
To be Published
|
|
6HGQ
 
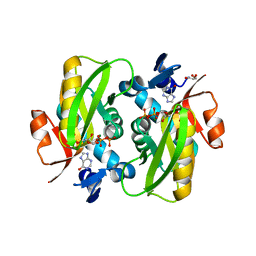 | | Crystal Structure of Human APRT wild type in complex with Hypoxanthine, PRPP and Mg2+ | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Adenine phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2018-08-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate selectivity and nucleophilic substitution mechanisms in human adenine phosphoribosyltransferase catalyzed reaction.
J.Biol.Chem., 294, 2019
|
|
6E11
 
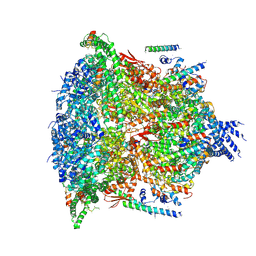 | | PTEX Core Complex in the Resetting (Compact) State | | Descriptor: | Endogenous cargo polypeptide, Exported protein 2, Heat shock protein 101, ... | | Authors: | Ho, C, Lai, M, Zhou, Z.H. | | Deposit date: | 2018-07-08 | | Release date: | 2018-08-22 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Malaria parasite translocon structure and mechanism of effector export.
Nature, 561, 2018
|
|
4N7C
 
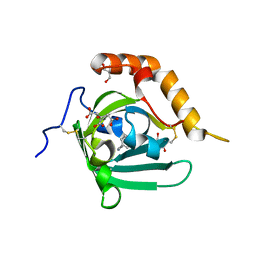 | | Structural re-examination of native Bla g 4 | | Descriptor: | 4-(2-aminoethyl)phenol, Bla g 4 allergen variant 1, CITRIC ACID, ... | | Authors: | Offermann, L.R, Chan, S.L, Osinski, T, Tan, Y.W, Chew, F.T, Sivaraman, J, Mok, Y.K, Minor, W, Chruszcz, M. | | Deposit date: | 2013-10-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The major cockroach allergen Bla g 4 binds tyramine and octopamine.
Mol.Immunol., 60, 2014
|
|
6TU3
 
 | | Rat 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Deshmukh, F.K, Polkinghorn, C.R, Elad, N, Sharon, M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Comparative Structural Analysis of 20S Proteasome Ortholog Protein Complexes by Native Mass Spectrometry.
Acs Cent.Sci., 6, 2020
|
|
1J2Q
 
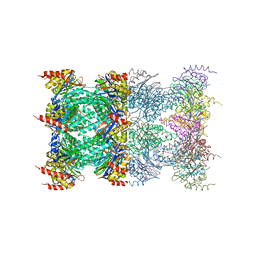 | | 20S proteasome in complex with calpain-Inhibitor I from archaeoglobus fulgidus | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID [1-(1-FORMYL-PENTYLCARBAMOYL)-3-METHYL-BUTYL]-AMIDE, Proteasome alpha subunit, Proteasome beta subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
4KWN
 
 | | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-24 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution
To be Published
|
|
6TR0
 
 | | Solution structure of U2AF2 RRM1,2 | | Descriptor: | Splicing factor U2AF 65 kDa subunit | | Authors: | Kang, H.-S, Sattler, M. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An autoinhibitory intramolecular interaction proof-reads RNA recognition by the essential splicing factor U2AF2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3RSG
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NAD. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3V1B
 
 | | Crystal structure of de novo designed MID1-apo2 | | Descriptor: | Computational design, MID1-apo2, GLYCEROL | | Authors: | Der, B.S, Machius, M, Miley, M.J, Kuhlman, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer.
J.Am.Chem.Soc., 134, 2012
|
|
4LD0
 
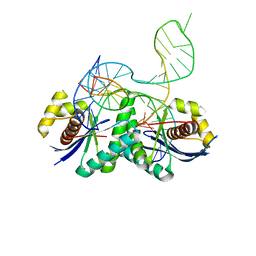 | | T. thermophilus RuvC in complex with Holliday junction substrate | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, DNA 11-MER, DNA 13-MER, ... | | Authors: | Gorecka, K.M, Komorowska, W, Nowotny, M. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Crystal structure of RuvC resolvase in complex with Holliday junction substrate.
Nucleic Acids Res., 41, 2013
|
|
6HL2
 
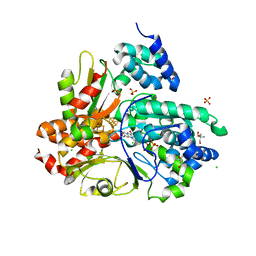 | | wild-type NuoEF from Aquifex aeolicus - oxidized form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
1J3R
 
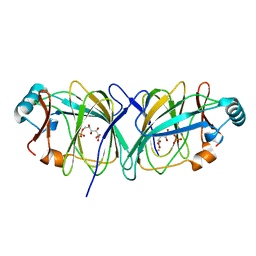 | | Crystal structure of Thermococcus litoralis phosphogrucose isomerase complexed with gluconate-6-phosphate | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, FE (III) ION, Phosphoglucose Isomerase | | Authors: | Jeong, J.-J, Fushinobu, S, Ito, S, Hidaka, M, Shoun, H, Wakagi, T. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of a novel cupin-type phosphoglucose isomerase
To be Published
|
|
6HLJ
 
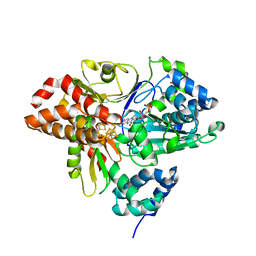 | | Variant G129S of NuoEF from Aquifex aeolicus - oxidized from | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
3RT9
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A | | Descriptor: | COENZYME A, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6E0E
 
 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
