3BHF
 
 | | Crystal structure of R49K mutant of Monomeric Sarcosine Oxidase crystallized in PEG as precipitant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Hassan-Abdallah, A, Zhao, G, Chen, Z, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2007-11-28 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arginine 49 is a bifunctional residue important in catalysis and biosynthesis of monomeric sarcosine oxidase: a context-sensitive model for the electrostatic impact of arginine to lysine mutations.
Biochemistry, 47, 2008
|
|
1L17
 
 | |
5INT
 
 | | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC | | Descriptor: | Phosphopantothenate--cysteine ligase | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
To Be Published
|
|
3B07
 
 | | Crystal structure of octameric pore form of gamma-hemolysin from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Yamashita, K, Kawai, Y, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Crystal structure of the octameric pore of
staphylococcal gamma-hemolysin reveals the beta-barrel
pore formation mechanism by two components
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5L1K
 
 | | PostInsertion complex of Human DNA Polymerase Eta bypassing an O6-Methyl-2'-deoxyguanosine : dC site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(6OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2'-deoxyguanosine by Human DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
3WK2
 
 | |
5L26
 
 | | Structure of CNTnw in an inward-facing substrate-bound state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease, SODIUM ION, ... | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
5L3Z
 
 | | polyketide ketoreductase SimC7 - binary complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, polyketide ketoreductase SimC7 | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-05 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
3WOI
 
 | | Crystal structure of the DAP BII (S657A) | | Descriptor: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2013-12-29 | | Release date: | 2014-09-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
5L45
 
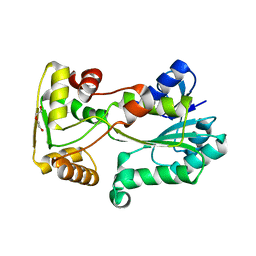 | | polyketide ketoreductase SimC7 - apo crystal form 2 | | Descriptor: | GLYCEROL, SimC7, TETRAETHYLENE GLYCOL | | Authors: | Schafer, M, Stevenson, C.E.M, Wilkinson, B, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-Assisted Catalysis in Polyketide Reduction Proceeds via a Phenolate Intermediate.
Cell Chem Biol, 23, 2016
|
|
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | Descriptor: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5L52
 
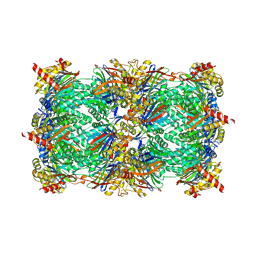 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 14 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
1JMS
 
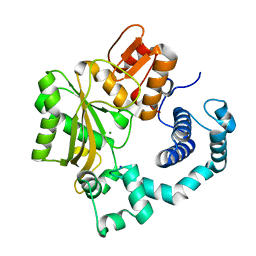 | | Crystal Structure of the Catalytic Core of Murine Terminal Deoxynucleotidyl Transferase | | Descriptor: | MAGNESIUM ION, SODIUM ION, TERMINAL DEOXYNUCLEOTIDYLTRANSFERASE | | Authors: | Delarue, M, Boule, J.B, Lescar, J, Expert-Bezancon, N, Sukumar, N, Jourdan, N, Rougeon, F, Papanicolaou, C. | | Deposit date: | 2001-07-19 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase.
Embo J., 21, 2002
|
|
5L5S
 
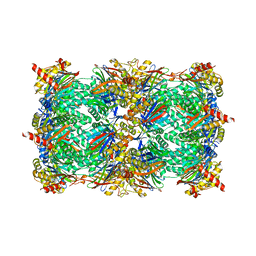 | |
5L55
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 18 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L6A
 
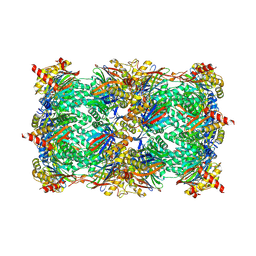 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 17 | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L5R
 
 | |
3WSX
 
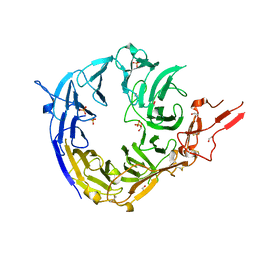 | | SorLA Vps10p domain in ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kitago, Y, Nakata, Z, Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2014-03-30 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for amyloidogenic peptide recognition by sorLA.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5L67
 
 | |
3WTH
 
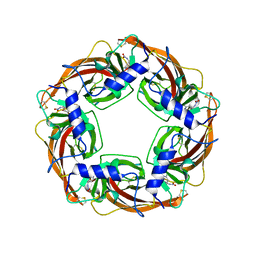 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
2ZPG
 
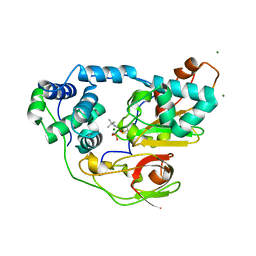 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 120min at 293K | | Descriptor: | FE (III) ION, MAGNESIUM ION, Nitrile hydratase subunit alpha, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
5L24
 
 | | Structure of CNTnw N149L in the intermediate 2 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
5L1J
 
 | |
2ZRR
 
 | | Crystal structure of an immunity protein that contributes to the self-protection of bacteriocin-producing Enterococcus mundtii 15-1A | | Descriptor: | Mundticin KS immunity protein | | Authors: | Jeon, H.J, Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2008-08-30 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mutagenic analysis of a bacteriocin immunity protein, Mun-im
Biochem.Biophys.Res.Commun., 378, 2009
|
|
2Z7X
 
 | | Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Lee, J.O, Jin, M.S, Kim, S.E, Heo, J.Y. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide
Cell(Cambridge,Mass.), 130, 2007
|
|
