5H9W
 
 | | Crystal structure of Regnase PIN domain, form II | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
1IEQ
 
 | | CRYSTAL STRUCTURE OF BARLEY BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, beta-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
6HY2
 
 | |
7P9W
 
 | | Epstein-Barr virus encoded apoptosis regulator BHRF1 in complex with Puma BH3 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, AMMONIUM ION, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.00010061 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
8QPZ
 
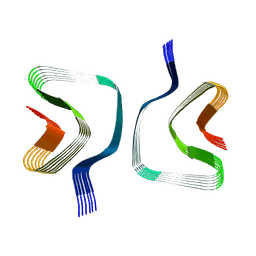 | | CryoEM structure of recombinant DeltaN7 alpha-synuclein in PBS | | Descriptor: | Alpha-synuclein | | Authors: | Thacker, D, Wilkinson, M, Dewison, K.M, Ranson, N.A, Brockwell, D.J, Radford, S.E. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Residues 2 to 7 of alpha-synuclein regulate amyloid formation via lipid-dependent and lipid-independent pathways.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8B46
 
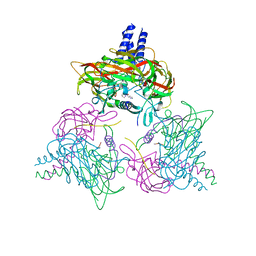 | | Crystal structure of the SUN1-KASH6 9:9 complex | | Descriptor: | CHLORIDE ION, Inositol 1,4,5-triphosphate receptor associated 2, POTASSIUM ION, ... | | Authors: | Gurusaran, M, Erlandsen, B.S, Davies, O.R. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The crystal structure of SUN1-KASH6 reveals an asymmetric LINC complex architecture compatible with nuclear membrane insertion.
Commun Biol, 7, 2024
|
|
8RFB
 
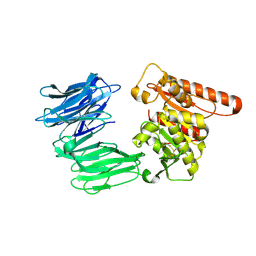 | | Cryo-EM structure of the R243C mutant of human Prolyl Endopeptidase-Like (PREPL) protein involved in Congenital myasthenic syndrome-22 (CMS22) | | Descriptor: | Prolyl endopeptidase-like | | Authors: | Theodoropoulou, A, Cavani, E, Antanasijevic, A, Marcaida, M.J, Dal Peraro, M. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Missense variants in CMS22 patients reveal that PREPL has both enzymatic and non-enzymatic functions.
JCI Insight, 2024
|
|
5HCW
 
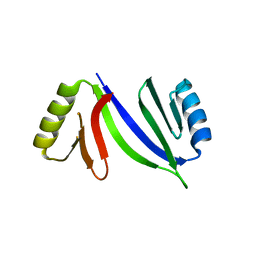 | |
5HFQ
 
 | | Crystal structure of the second bromodomain Q443H mutant of human BRD2 | | Descriptor: | Bromodomain-containing protein 2 | | Authors: | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Fonseca, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the second bromodomain Q443H mutant of human BRD2
To Be Published
|
|
2YN2
 
 | | Huf protein - paralogue of the tau55 histidine phosphatase domain | | Descriptor: | FORMIC ACID, UNCHARACTERIZED PROTEIN YNL108C | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Tfiiic.
J.Biol.Chem., 288, 2013
|
|
5HEV
 
 | |
1IN1
 
 | |
5HEZ
 
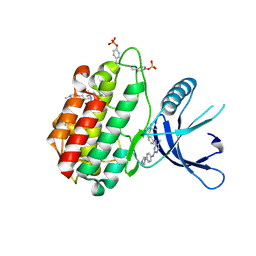 | | JAK2 kinase (JH1 domain) mutant P1057A in complex with TG101209 | | Descriptor: | CHLORIDE ION, N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide, Tyrosine-protein kinase JAK2, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity.
Sci Transl Med, 8, 2016
|
|
5OY2
 
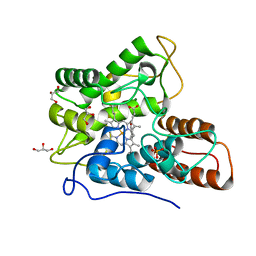 | | Direct-evolutioned unspecific peroxygenase from Agrocybe aegerita, in complex with DMP | | Descriptor: | 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2017-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Insights into the Substrate Promiscuity of a Laboratory-Evolved Peroxygenase.
Acs Chem.Biol., 13, 2018
|
|
5DTZ
 
 | |
5DZ8
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA variable (V) domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BspA (BspA_V), ... | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
5HFK
 
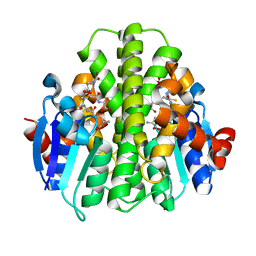 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
3L84
 
 | | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | ACETATE ION, GLYCEROL, Transketolase | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-29 | | Release date: | 2010-02-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
5DUQ
 
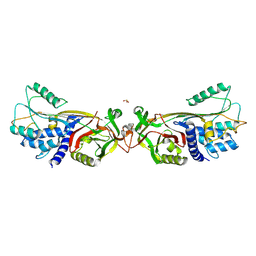 | | Active human c1-inhibitor in complex with dextran sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma protease C1 inhibitor, SULFITE ION, ... | | Authors: | Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A, Pannu, N.S. | | Deposit date: | 2015-09-20 | | Release date: | 2016-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
8QDJ
 
 | |
1INY
 
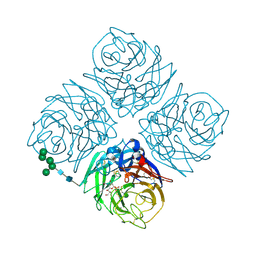 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
5HGI
 
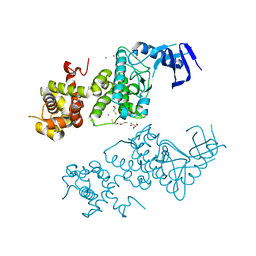 | | Crystal structure of apo human IRE1 alpha | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CESIUM ION, ... | | Authors: | Feldman, H.C, Tong, M, Wang, L, Meza-Acevedo, R, Gobillot, T.A, Gliedt, J.M, Hari, S.B, Mitra, A.K, Backes, B.J, Papa, F.R, Seeliger, M.A, Maly, D.J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Inhibition of IRE1 alpha with ATP-Competitive Ligands.
Acs Chem.Biol., 11, 2016
|
|
5SB8
 
 | | Tubulin-maytansinoid-3-complex | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R)-11-chloro-6-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18,21-hexaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Marzullo, P, Boiarska, Z, Perez-Pena, H, Abel, A.-C, Alvarez-Bernad, B, Lucena-Agell, D, Vasile, F, Sironi, M, Steinmetz, M.O, Prota, A.E, Diaz, J.F, Pieraccini, S, Passarella, D. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Maytansinol Derivatives: Side Reactions as a Chance for New Tubulin Binders.
Chemistry, 28, 2022
|
|
5E2C
 
 | | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Chang, C, Endres, L, Endres, M, SACCHETTINI, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
5HGR
 
 | | Structure of Anabaena (Nostoc) sp. PCC 7120 Orange Carotenoid Protein binding canthaxanthin | | Descriptor: | Orange Carotenoid Protein (OCP), beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Leverenz, R.L, Kerfeld, C.A. | | Deposit date: | 2016-01-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Different Functions of the Paralogs to the N-Terminal Domain of the Orange Carotenoid Protein in the Cyanobacterium Anabaena sp. PCC 7120.
Plant Physiol., 171, 2016
|
|
