6ES4
 
 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
6ESD
 
 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A semi-synthetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2017
|
|
8DFW
 
 | | Crystal Structure of Human BTN2A1 in Complex With Vgamma9-Vdelta2 T Cell Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
6YWA
 
 | |
5ON6
 
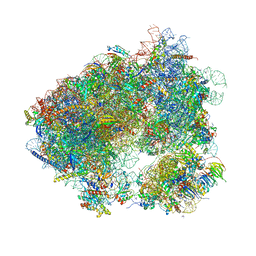 | | Crystal structure of haemanthamine bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Meyer, M, Yusupova, G, Yusupov, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.10000229 Å) | | Cite: | The Amaryllidaceae Alkaloid Haemanthamine Binds the Eukaryotic Ribosome to Repress Cancer Cell Growth.
Structure, 26, 2018
|
|
6ECD
 
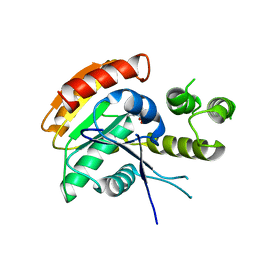 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a tetradepsipeptide | | Descriptor: | Vlm2, tetradepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
8D40
 
 | |
6Z61
 
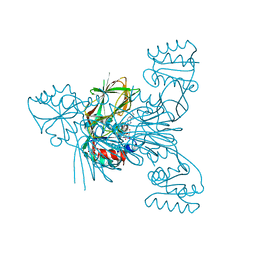 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-[6-azanyl-8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-[6-(2-azanylethylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]purin-9-yl]-5-(hydroxymethyl)oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
6Z65
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | CITRIC ACID, NAD kinase 1, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-4-azanyl-butanamide | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
8CQL
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-N-((S)-1-(5-Fluoro-2-methoxy-4-(4-methylthiazol-5-yl)phenyl)ethyl)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamide (Compound 33) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[5-fluoranyl-2-methoxy-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
6EHH
 
 | | Crystal structure of mouse MTH1 mutant L116M with inhibitor TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gustafsson, R, Narwal, M, Jemth, A.-S, Almlof, I, Warpman Berglund, U, Helleday, T, Stenmark, P. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
8CQK
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-Fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(2-methyl-4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (Compound 30) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-[2-methyl-4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
8CPY
 
 | |
6ZGK
 
 | | GLIC pentameric ligand-gated ion channel, pH 3 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6E5D
 
 | |
6E5L
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-20 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
8CQE
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-Fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(2-methyl-4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (Compound 37) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-7-fluoranyl-6-(4-methyl-1,3-thiazol-5-yl)-1,2,3,4-tetrahydronaphthalen-1-yl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Casement, R, Phuong Vu, L, Ciulli, A, Gutschow, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Expanding the Structural Diversity at the Phenylene Core of Ligands for the von Hippel-Lindau E3 Ubiquitin Ligase: Development of Highly Potent Hypoxia-Inducible Factor-1 alpha Stabilizers.
J.Med.Chem., 66, 2023
|
|
6ZGD
 
 | | GLIC pentameric ligand-gated ion channel, pH 7 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6E5S
 
 | |
6ZGJ
 
 | | GLIC pentameric ligand-gated ion channel, pH 5 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6Z13
 
 | | VEGF-A 13:107 crystallized with 3C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, AMINO GROUP, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
6Z64
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]prop-2-ynyl-(3-azanylpropyl)amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2020-05-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | New Chemical Probe Targeting Bacterial NAD Kinase.
Molecules, 25, 2020
|
|
6Z3F
 
 | | VEGF-A 13:107 crystallized with 2C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, AMINO GROUP, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
5OVY
 
 | | Crystal structure of MAB_4384 tetR | | Descriptor: | Putative transcriptional regulator, TetR family | | Authors: | Richard, M, Gutierrez, A.V, Viljoen, A, Ghigo, E, Blaise, M, Kremer, L. | | Deposit date: | 2017-08-30 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Structural Insights Into the Unique TetR-Dependent Regulation of a Drug Efflux Pump inMycobacterium abscessus.
Front Microbiol, 9, 2018
|
|
5JD2
 
 | | SFX structure of corestreptavidin-selenobiotin complex | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-selenopheno[3,4-d]imidazol-4-yl]pentanoic acid, Streptavidin | | Authors: | DeMirci, H, Hunter, M.S, Boutet, S. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser.
Nat Commun, 7, 2016
|
|
