4N2I
 
 | | Crystal structure of Protein Arginine Deiminase 2 (D177A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5X12
 
 | | Crystal structure of Bacillus subtilis PadR | | Descriptor: | Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
8J2Q
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5X2G
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5W8W
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 - new refinement | | Descriptor: | Metallo-beta-lactamase type 2, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Gonzalez, J.M, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Wlodawer, A, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
8INX
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor ensitrelvir
To Be Published
|
|
8IP2
 
 | | Escherichia coli OpgG mutant-D361N with beta-1,2-glucan | | Descriptor: | Glucans biosynthesis protein G, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Motouchi, S, Nakajima, M. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-13 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification of enzymatic functions of osmo-regulated periplasmic glucan biosynthesis proteins from Escherichia coli reveals a novel glycoside hydrolase family.
Commun Biol, 6, 2023
|
|
3ZHB
 
 | |
4MLQ
 
 | | Crystal structure of Bacillus megaterium porphobilinogen deaminase | | Descriptor: | 3-[(5S)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, ACETIC ACID, ... | | Authors: | Azim, N, Deery, E, Warren, M.J, Erskine, P, Cooper, J.B, Coker, A, Wood, S.P, Akhtar, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for the partially oxidized dipyrromethene and dipyrromethanone forms of the cofactor of porphobilinogen deaminase: structures of the Bacillus megaterium enzyme at near-atomic resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8JH8
 
 | | Structure-based characterization and improvement of an enzymatic activity of Acremonium alcalophilum feruloyl esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Feruloyl esterase, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Characterization and Improvement of an Enzymatic Activity of Acremonium alcalophilum Feruloyl Esterase
Acs Sustain Chem Eng, 12, 2024
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WQ8
 
 | |
8IP0
 
 | | Cryo-EM structure of type I-B Cascade bound to a PAM-containing dsDNA target at 3.6 angstrom resolution | | Descriptor: | CRISPR associated protein Cas11b, DNA (41-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and genome editing of type I-B CRISPR-Cas.
Nat Commun, 15, 2024
|
|
4MQ3
 
 | |
8JH9
 
 | | Structure-based characterization and improvement of an enzymatic activity of Acremonium alcalophilum feruloyl esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Feruloyl esterase with ferulic acid, ... | | Authors: | Phienluphon, A, Kondo, K, Mikami, B, Nagata, T, Katahira, M. | | Deposit date: | 2023-05-22 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Characterization and Improvement of an Enzymatic Activity of Acremonium alcalophilum Feruloyl Esterase
Acs Sustain Chem Eng, 12, 2024
|
|
4MQL
 
 | | Crystal structure of Antigen 85C-C209S mutant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
5WXQ
 
 | |
3ZXI
 
 | | Crystal structure of human mitochondrial tyrosyl-tRNA synthetase in complex with a tyrosyl-adenylate analog | | Descriptor: | PHOSPHORIC ACID 2-AMINO-3-(4-HYDROXY-PHENYL)-PROPYL ESTER ADENOSIN-5'YL ESTER, TYROSYL-TRNA SYNTHETASE, MITOCHONDRIAL | | Authors: | Bonnefond, L, Frugier, M, Rudinger-Thirion, J, Balg, C, Chenevert, R, Lorber, B, Giege, R, Sauter, C. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
5WY9
 
 | |
4MTV
 
 | | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Shukla, P.K, Chaudhary, A, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling Glycoprotein with pentasaccharide at 2.8A resolution
To be Published
|
|
5X0F
 
 | |
8J7C
 
 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.88A with an arsenic ion bound at Cys57 | | Descriptor: | ARSENIC, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Attarataya, J, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Mungthin, M, Leelayoova, S, Choowongkomon, K, Leartsakulpanich, U. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
5X11
 
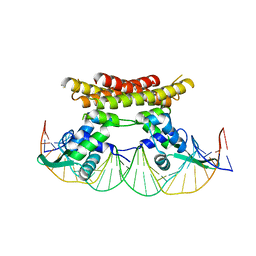 | | Crystal structure of Bacillus subtilis PadR in complex with operator DNA | | Descriptor: | DNA (28-MER), Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
4MV0
 
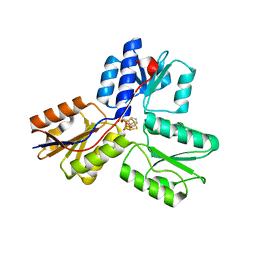 | | IspH in complex with pyridin-2-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-2-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
4MX8
 
 | |
