6X0N
 
 | |
5N0Z
 
 | | hPAD4 crystal complex with AFM-41a | | Descriptor: | 2-ethyl-~{N}-[(1~{S})-4-(2-fluoranylethanimidoylamino)-1-(4-methoxy-1-methyl-benzimidazol-2-yl)butyl]-3-oxidanylidene-1~{H}-isoindole-4-carboxamide, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Beaumont, E, Kerry, P, Thompson, P, Muth, A, Subramanian, V, Nagar, M, Srinath, H, Clancy, K, Parelkar, S. | | Deposit date: | 2017-02-03 | | Release date: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Development of a Selective Inhibitor of Protein Arginine Deiminase 2.
J. Med. Chem., 60, 2017
|
|
6WYL
 
 | | Cryo-EM structure of GltPh L152C-G351C mutant in the intermediate outward-facing state. | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
5N1B
 
 | | hPAD4 crystal complex with AFM-14a | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, SULFATE ION, ... | | Authors: | Beaumont, E, Kerry, P, Thompson, P, Muth, A, Subramanian, V, Nagar, M, Srinath, H, Clancy, K, Parelkar, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-24 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of a Selective Inhibitor of Protein Arginine Deiminase 2.
J. Med. Chem., 60, 2017
|
|
6WYK
 
 | | Cryo-EM structure of the GltPh L152C-G321C mutant in the intermediate chloride conducting state. | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
5N4J
 
 | |
5NB8
 
 | | Structure of vWC domain from CCN3 | | Descriptor: | GLYCEROL, IMIDAZOLE, Protein NOV homolog | | Authors: | Xu, E.-R, Hyvonen, M. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analyses of von Willebrand factor C domains of collagen 2A and CCN3 reveal an alternative mode of binding to bone morphogenetic protein-2.
J. Biol. Chem., 292, 2017
|
|
4TV8
 
 | | Tubulin-Maytansine complex | | Descriptor: | (3beta,4beta,5beta,10beta,11E,13E)-maytansine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Bargsten, K, Diaz, J.F, Marsh, M, Cuevas, C, Liniger, M, Neuhaus, C, Andreu, J.M, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5NCQ
 
 | | Structure of the (SR) Ca2+-ATPase bound to a Tetrahydrocarbazole and TNP-ATP | | Descriptor: | (1~{S})-~{N}-[(4-bromophenyl)methyl]-7-(trifluoromethyloxy)-2,3,4,9-tetrahydro-1~{H}-carbazol-1-amine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, ... | | Authors: | Bublitz, M, Kjellerup, L, O'Hanlon Cohrt, K, Gordon, S, Mortensen, A.L, Clausen, J.D, Pallin, D, Hansen, J.B, Brown, W.D, Fuglsang, A, Winther, A.-M.L. | | Deposit date: | 2017-03-06 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetrahydrocarbazoles are a novel class of potent P-type ATPase inhibitors with antifungal activity.
PLoS ONE, 13, 2018
|
|
5ND8
 
 | | Hibernating ribosome from Staphylococcus aureus (Unrotated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Ayupov, R, Usachev, K, Myasnikov, A, Simonetti, A, Validov, S, Kieffer, B, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
7QZX
 
 | | Human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide | | Descriptor: | 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carboxamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7R1X
 
 | | human Carbonic Anhydrase II in complex with 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide | | Descriptor: | 4-oxo-N-(4-sulfamoylphenethyl)-1,3,4,6,7,11b-hexahydro-2H-pyrazino[2,1-a]isoquinoline-2-carbothioamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Development of Praziquantel sulphonamide derivatives as antischistosomal drugs.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7R2S
 
 | | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas
To Be Published
|
|
6WH0
 
 | | Crystal structure of HyBcl-2-4 with HyBax BH3 | | Descriptor: | Apoptosis regulator BAX, Maltodextrin-binding protein,Bcl-2-like 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kvansakul, M, Hinds, M.G, Banjara, S. | | Deposit date: | 2020-04-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structural basis of Bcl-2 mediated cell death regulation in hydra.
Biochem.J., 477, 2020
|
|
7R2P
 
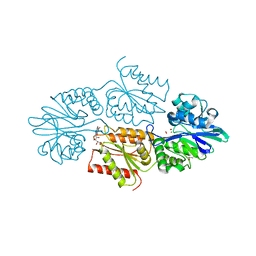 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
6WO6
 
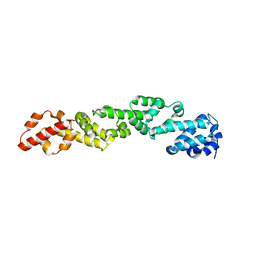 | |
7R1J
 
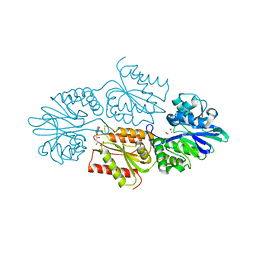 | | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
7R0F
 
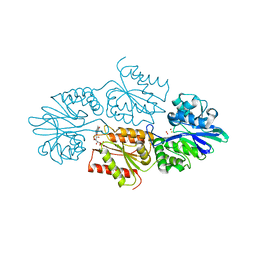 | | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R2O
 
 | | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R2R
 
 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
7R1H
 
 | | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli
To Be Published
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NIS
 
 | |
5NF3
 
 | | The fimbrial shaft protein Mfa1 from Porphyromonas gingivalis-C-terminal deletion | | Descriptor: | ACETATE ION, CALCIUM ION, Minor fimbrium subunit Mfa1 | | Authors: | Hall, M, Hasegawa, Y, Persson, K, Yoshimura, F. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterization of shaft, anchor, and tip proteins of the Mfa1 fimbria from the periodontal pathogen Porphyromonas gingivalis.
Sci Rep, 8, 2018
|
|
5NJA
 
 | | E. coli Microcin-processing metalloprotease TldD/E with angiotensin analogue bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HIS-PRO-PHE, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
