5OO7
 
 | |
6T5I
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with inhibitor of WNT production (IWP)-2 | | Descriptor: | DIMETHYL SULFOXIDE, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Oelker, M, Grundstrom, C, Blumenthal, A, Sauer-Eriksson, A.E. | | Deposit date: | 2019-10-16 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of the master regulator of Listeria monocytogenes virulence enables bacterial clearance from spacious replication vacuoles in infected macrophages.
Plos Pathog., 18, 2022
|
|
4MXI
 
 | | ClpP Ser98dhA | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Gersch, M, Kolb, R, Alte, F, Groll, M, Sieber, S.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of Oligomerization and Dehydroalanine Formation as Mechanisms for ClpP Protease Inhibition.
J.Am.Chem.Soc., 136, 2014
|
|
4MZ1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 | | Descriptor: | 1-(4-bromophenyl)-3-{2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}urea, ACETIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3991 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12
To be Published, 2013
|
|
6TO6
 
 | | Solution structure of the modulator of repression (MOR) of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | MOR | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Lo Leggio, L, Jensen, M.R. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2AIB
 
 | | beta-cinnamomin in complex with ergosterol | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-elicitin cinnamomin, ERGOSTEROL, ... | | Authors: | Rodrigues, M.L, Archer, M, Martel, P, Miranda, S, Thomaz, M, Enguita, F.J, Baptista, R.P, Melo, E.P, Sousa, N, Cravador, A, Carrondo, M.A. | | Deposit date: | 2005-07-29 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of the free and sterol-bound forms of beta-cinnamomin
Biochim.Biophys.Acta, 1764, 2006
|
|
1ST4
 
 | | Structure of DcpS bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
5OVJ
 
 | | Structure of the apo form of Mycobacterium smegmatis MabA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, SULFATE ION | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4L6E
 
 | | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2. Northeast Structural Genomics Consortium (NESG) Target HR9193b | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Kogan, S, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-12 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2.
To be Published
|
|
5OSX
 
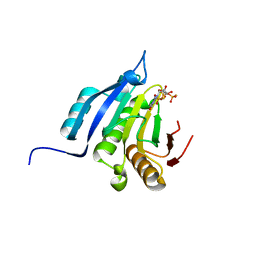 | | Translation initiation factor 4E in complex with m7G(5'S)ppp(5'S)G mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, POTASSIUM ION, [(2~{S},3~{S},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanylidene-3~{H}-purin-7-ium-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-[[[(3~{R},4~{S})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-phosphinic acid | | Authors: | Warminski, M, Nowak, E, Wojtczak, B.A, Fac-Dabrowska, K, Kubacka, D, Nowicka, A, Sikorski, P.J, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 5'-Phosphorothiolate Dinucleotide Cap Analogues: Reagents for Messenger RNA Modification and Potent Small-Molecular Inhibitors of Decapping Enzymes.
J. Am. Chem. Soc., 140, 2018
|
|
4MUA
 
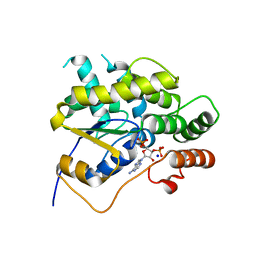 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, Sulfotransferase | | Authors: | Valentim, C.L.L, Cioli, D, Chevalier, F.D, Cao, X, Taylor, A.B, Holloway, S.P, Pica-Mattoccia, L, Guidi, A, Basso, A, Tsai, I.J, Berriman, M, Carvalho-Queiroz, C, Almeida, M, Aguilar, H, Frantz, D.E, Hart, P.J, Anderson, T.J.C, LoVerde, P.T. | | Deposit date: | 2013-09-21 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetic and molecular basis of drug resistance and species-specific drug action in schistosome parasites.
Science, 342, 2013
|
|
2V1M
 
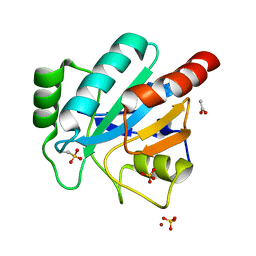 | | Crystal structure of Schistosoma mansoni glutathione peroxidase | | Descriptor: | ACETATE ION, GLUTATHIONE PEROXIDASE, LITHIUM ION, ... | | Authors: | Dimastrogiovanni, D, Miele, A.E, Angelucci, F, Boumis, G, Bellelli, A, Brunori, M. | | Deposit date: | 2008-09-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Combining Crystallography and Molecular Dynamics: The Case of Schistosoma Mansoni Phospholipid Glutathione Peroxidase.
Proteins, 78, 2010
|
|
2VBT
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and PO4 | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
1JDE
 
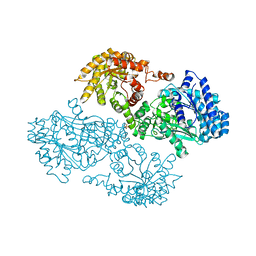 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
1T4N
 
 | | Solution structure of Rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
2UYO
 
 | | Crystal structure of ML2640c from Mycobacterium leprae in an hexagonal crystal form | | Descriptor: | HYPOTHETICAL PROTEIN ML2640 | | Authors: | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Miras, I, Shepard, W, Alzari, P.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
1B5V
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
4LJI
 
 | | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, Cytochrome c-550-like protein, HEME C | | Authors: | Suga, M, Lai, T.-L, Sugiura, M, Shen, J.-R, Boussac, A. | | Deposit date: | 2013-07-04 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Crystal structure at 1.5 angstrom resolution of the PsbV2 cytochrome from the cyanobacterium Thermosynechococcus elongatus
Febs Lett., 587, 2013
|
|
2V8C
 
 | | Mouse Profilin IIa in complex with the proline-rich domain of VASP | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
5OML
 
 | | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope | | Descriptor: | (3~{R})-3-[[1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]carbonylamino]-3-phenyl-propanoic acid, BETA-MERCAPTOETHANOL, Peroxin 14, ... | | Authors: | Ratkova, E.L, Dawidowski, M, Napolitano, V, Dubin, G, Fino, R, Popowicz, G, Sattler, M, Tetko, I.V. | | Deposit date: | 2017-08-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope
To Be Published
|
|
4MO1
 
 | | Crystal structure of antitermination protein Q from bacteriophage lambda. Northeast Structural Genomics Consortium target OR18A. | | Descriptor: | Antitermination protein Q, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Vorobiev, S, Su, M, Nickels, B, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Ebright, R.H, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of antitermination protein Q from bacteriophage lambda.
To be Published
|
|
5ZJY
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-LYS-ARG-TYR-SER-ARG-2JN-GLN-LEU-LEU-2JN-PHE | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, B, Uhring, M. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZK5
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-ARG-TYR-SER-ARG-GLU-GLN-LEU-LEU-MK8-PHE-GLN-ARG-MK8 | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
1R42
 
 | | Native Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ZINC ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
