2CLH
 
 | | Tryptophan Synthase in complex with (naphthalene-2'-sulfonyl)-2-amino- 1-ethylphosphate (F19) | | Descriptor: | 2-[(2-NAPHTHYLSULFONYL)AMINO]ETHYL DIHYDROGEN PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and characterization of allosteric probes of substrate channeling in the tryptophan synthase bienzyme complex.
Biochemistry, 46, 2007
|
|
7N2U
 
 | | Elongating 70S ribosome complex in a hybrid-H1 pre-translocation (PRE-H1) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
2CT5
 
 | | Solution Structure of the zinc finger BED domain of the zinc finger BED domain containing protein 1 | | Descriptor: | ZINC ION, Zinc finger BED domain containing protein 1 | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the zinc finger BED domain of the zinc finger BED domain containing protein 1
To be Published
|
|
2CTR
 
 | | Solution structure of J-domain from human DnaJ subfamily B menber 9 | | Descriptor: | DnaJ homolog subfamily B member 9 | | Authors: | Kobayashi, N, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-24 | | Release date: | 2005-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of J-domain from human DnaJ subfamily B menber 9
To be Published
|
|
2CNM
 
 | | RimI - Ribosomal S18 N-alpha-protein acetyltransferase in complex with a bisubstrate inhibitor (Cterm-Arg-Arg-Phe-Tyr-Arg-Ala-N-alpha- acetyl-S-CoA). | | Descriptor: | 30S RIBOSOMAL PROTEIN S18, COENZYME A, MODIFICATION OF 30S RIBOSOMAL SUBUNIT PROTEIN S18 | | Authors: | Vetting, M.W, Yu, M, Bareich, D.C, Blanchard, J.S. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Rimi from Salmonella Typhimurium Lt2, the Gnat Responsible for N{Alpha}- Acetylation of Ribosomal Protein S18.
Protein Sci., 17, 2008
|
|
2COF
 
 | | Solution structure of the C-terminal PH domain of hypothetical protein KIAA1914 from human | | Descriptor: | Protein KIAA1914 | | Authors: | Li, H, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal PH domain of hypothetical protein KIAA1914 from human
To be Published
|
|
2COZ
 
 | | Solution structure of the CAP-Gly domain in human centrosome-associated protein CAP350 | | Descriptor: | centrosome-associated protein 350 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CAP-Gly domain in human centrosome-associated protein CAP350
To be Published
|
|
2CV9
 
 | |
2CRU
 
 | | Solution structure of programmed cell death 5 | | Descriptor: | Programmed cell death protein 5 | | Authors: | Nagashima, T, Izumi, K, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of programmed cell death 5
To be published
|
|
2CWF
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
2CTD
 
 | | Solution structure of two zf-C2H2 domains from human Zinc finger protein 512 | | Descriptor: | ZINC ION, Zinc finger protein 512 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-24 | | Release date: | 2005-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of two zf-C2H2 domains from human Zinc finger protein 512
To be Published
|
|
2CWW
 
 | | Crystal structure of Thermus thermophilus TTHA1280, a putative SAM-dependent RNA methyltransferase, in complex with S-adenosyl-L-homocysteine | | Descriptor: | ACETIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7N1P
 
 | | Elongating 70S ribosome complex in a classical pre-translocation (PRE-C) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, S.K, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
4ARB
 
 | | Mus musculus Acetylcholinesterase in complex with (S)-C5685 at 2.25 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ACETYLCHOLINESTERASE, ... | | Authors: | Berg, L, Niemiec, M.S, Qian, W, Andersson, C.D, WittungStafshede, P, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-04-23 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Similar But Different: Thermodynamic and Structural Characterization of a Pair of Enantiomers Binding to Acetylcholinesterase.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2CXE
 
 | | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal) | | Descriptor: | Ribulose bisphosphate carboxylase | | Authors: | Mizohata, E, Mishima, C, Akasaka, R, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of octameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) from Pyrococcus horikoshii OT3 (form-2 crystal)
To be Published
|
|
2CZ1
 
 | | photo-activation state of Fe-type NHase with n-BA in anaerobic condition | | Descriptor: | BUTANOIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Kawano, Y, Hashimoto, K, Odaka, M, Nakayama, H, Takio, K, Endo, I, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-09 | | Release date: | 2006-01-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | photo-activation state of Fe-type NHase with n-BA in anaerobic condition
To be Published
|
|
2CV4
 
 | | Crystal Structure of an Archaeal Peroxiredoxin from the Aerobic Hyperthermophilic Crenarchaeon Aeropyrum pernix K1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ISOPROPYL ALCOHOL, peroxiredoxin | | Authors: | Mizohata, E, Sakai, H, Fusatomi, E, Terada, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-31 | | Release date: | 2005-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Archaeal Peroxiredoxin from the Aerobic Hyperthermophilic Crenarchaeon Aeropyrum pernix K1
J.Mol.Biol., 354, 2005
|
|
2A7T
 
 | | Crystal Structure of a novel neurotoxin from Buthus tamalus at 2.2A resolution. | | Descriptor: | Neurotoxin | | Authors: | Ethayathulla, A.S, Sharma, M, Saravanan, K, Sharma, S, Kaur, P, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-07-06 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a highly acidic neurotoxin from scorpion Buthus tamulus at 2.2A resolution reveals novel structural features.
J.Struct.Biol., 155, 2006
|
|
7O2L
 
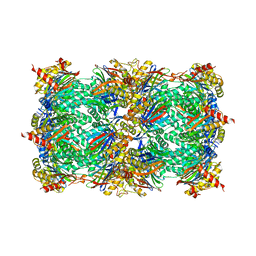 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
5HD3
 
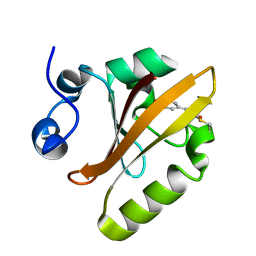 | |
2ADE
 
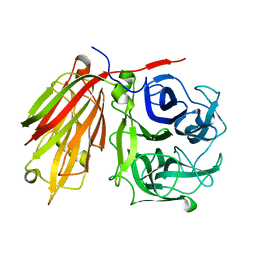 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus in complex with fructose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-fructofuranose, ... | | Authors: | Verhaest, M, Le Roy, K, De Ranter, C.J, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor.
New Phytol, 174, 2007
|
|
2AEG
 
 | | X-Ray Crystal Structure of Protein Atu5096 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR63. | | Descriptor: | hypothetical protein AGR_pAT_140 | | Authors: | Forouhar, F, Abashidze, M, Kuzin, A.P, Vorobiev, S.M, Shastry, R, Cooper, B, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Hypothetical Protein Atu5096 from Agrobacterium tumefaciens.
To be Published
|
|
1R4N
 
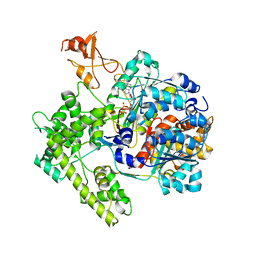 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ubiquitin-like protein NEDD8, ZINC ION, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
2A14
 
 | | Crystal Structure of Human Indolethylamine N-methyltransferase with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, indolethylamine N-methyltransferase | | Authors: | Dong, A, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Human Indolethylamine
N-methyltransferase in complex with SAH.
To be Published
|
|
2A43
 
 | | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif | | Descriptor: | MAGNESIUM ION, RNA Pseudoknot | | Authors: | Pallan, P.S, Marshall, W.S, Harp, J, Jewett III, F.C, Wawrzak, Z, Brown II, B.A, Rich, A, Egli, M. | | Deposit date: | 2005-06-27 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Biochemistry, 44, 2005
|
|
