1Y52
 
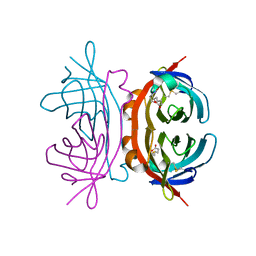 | | structure of insect cell (Baculovirus) expressed AVR4 (C122S)-biotin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin-related protein 4/5, BIOTIN | | Authors: | Eisenberg-Domovich, Y, Hytonen, V.P, Wilchek, M, Bayer, E.A, Kulomaa, M.S, Livnah, O. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-24 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structure of an avidin-related protein: insight into high-affinity biotin binding and protein stability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4J5P
 
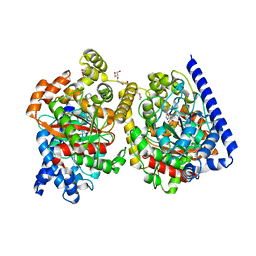 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (1S)-1-{5-[5-(bromomethyl)pyridin-2-yl]-1,3-oxazol-2-yl}-7-phenylheptan-1-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otrubova, K, Brown, M, McCormick, M.S, Han, G.W, O'Neal, S.T, Cravatt, B.F, Stevens, R.C, Lichtman, A.H, Boger, D.L. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design of Fatty Acid amide hydrolase inhibitors that act by covalently bonding to two active site residues.
J.Am.Chem.Soc., 135, 2013
|
|
1Q5Z
 
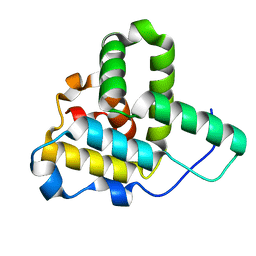 | | Crystal Structure of the C-terminal Actin Binding Domain of Salmonella Invasion Protein A (SipA) | | Descriptor: | SipA | | Authors: | Stebbins, C.E, Lilic, M, Galkin, V.E, Orlova, A, VanLoock, M.S, Egelman, E.H. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms.
Science, 301, 2003
|
|
1PWY
 
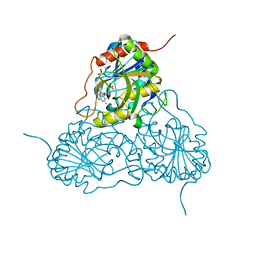 | | CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH ACYCLOVIR | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Dos Santos, D.M, Canduri, F, Pereira, J.H, Vinicius Bertacine Dias, M, Silva, R.G, Mendes, M.A, Palma, M.S, Basso, L.A, De Azevedo, W.F, Santos, D.S. | | Deposit date: | 2003-07-02 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human purine nucleoside phosphorylase complexed with acyclovir.
Biochem.Biophys.Res.Commun., 308, 2003
|
|
4M92
 
 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
 | | crystal structure of hN33/Tusc3-peptide 1 | | Descriptor: | Protein cereblon, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M32
 
 | | Crystal structure of gated-pore mutant D138N of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
1MFM
 
 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
4M33
 
 | | Crystal structure of gated-pore mutant H141D of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
1NAQ
 
 | | Crystal structure of CUTA1 from E.coli at 1.7 A resolution | | Descriptor: | MERCURIBENZOIC ACID, MERCURY (II) ION, Periplasmic divalent cation tolerance protein cutA | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Viezzoli, M.S, Banci, L, Bertini, I, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-11-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction.
J.Biol.Chem., 278, 2003
|
|
1W36
 
 | | RecBCD:DNA complex | | Descriptor: | CALCIUM ION, DNA HAIRPIN, EXODEOXYRIBONUCLEASE V ALPHA CHAIN, ... | | Authors: | Singleton, M.R, Dillingham, M.S, Gaudier, M, C Kowalczykowski, S, Wigley, D.B. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Recbcd Enzyme Reveals a Machine for Processing DNA Breaks
Nature, 432, 2004
|
|
2JXR
 
 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
1QG2
 
 | | CANINE GDP-RAN R76E MUTANT | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RAN) | | Authors: | Kent, H.M, Moore, M.S, Quimby, B.B, Baker, A.M.E, McCoy, A.J, Murphy, G.A, Corbett, A.H, Stewart, M. | | Deposit date: | 1999-04-20 | | Release date: | 1999-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered mutants in the switch II loop of Ran define the contribution made by key residues to the interaction with nuclear transport factor 2 (NTF2) and the role of this interaction in nuclear protein import.
J.Mol.Biol., 289, 1999
|
|
4MBQ
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4M35
 
 | | Crystal structure of gated-pore mutant H126/141D of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
4M34
 
 | | Crystal structure of gated-pore mutant D138H of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
1QG4
 
 | | CANINE GDP-RAN F72Y MUTANT | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RAN) | | Authors: | Kent, H.M, Moore, M.S, Quimby, B.B, Baker, A.M.E, McCoy, A.J, Murphy, G.A, Corbett, A.H, Stewart, M. | | Deposit date: | 1999-04-20 | | Release date: | 1999-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered mutants in the switch II loop of Ran define the contribution made by key residues to the interaction with nuclear transport factor 2 (NTF2) and the role of this interaction in nuclear protein import.
J.Mol.Biol., 289, 1999
|
|
1QJT
 
 | | SOLUTION STRUCTURE OF THE APO EH1 DOMAIN OF MOUSE EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE 15, EPS15 | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE SUBSTRATE 15, EPS15 | | Authors: | Whitehead, B, Tessari, M, Carotenuto, A, van Bergen en Henegouwen, P.M, Vuister, G.W. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eh1 Domain of Eps15 is Structurally Classified as a Member of the S100 Subclass of EF-Hand Containing Proteins
Biochemistry, 38, 1999
|
|
1QD5
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Ubarretxena-Belandia, I, Blaauw, M, Kalk, K.H, Verheij, H.M, Egmond, M.R, Dekker, N, Dijkstra, B.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural evidence for dimerization-regulated activation of an integral membrane phospholipase.
Nature, 401, 1999
|
|
6ZJ3
 
 | | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Matzov, D, Halfon, H, Zimmerman, E, Rozenberg, H, Bashan, A, Gray, M.W, Yonath, A.E, Shalev-Benami, M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis.
Nucleic Acids Res., 48, 2020
|
|
3ZN2
 
 | | protein engineering of halohydrin dehalogenase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, HALOHYDRIN DEHALOGENASE, ... | | Authors: | Schallmey, M, Jekel, P, Tang, L, Majeric-Elenkov, M, Hoeffken, H.W, Hauer, B, Janssen, D.B. | | Deposit date: | 2013-02-13 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Single Point Mutation Enhances Hydroxynitrile Synthesis by Halohydrin Dehalogenase.
Enzyme.Microb.Technol., 70, 2015
|
|
5WVX
 
 | | Crystal Structure of bifunctional Kunitz type Trypsin /amylase inhibitor (AMTIN) from the tubers of Alocasia macrorrhiza | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CITRIC ACID, Trypsin/chymotrypsin inhibitor | | Authors: | Palayam, M, Radhakrishnan, M, Lakshminarayanan, K, Balu, K.E, Ganapathy, J, Krishnasamy, G. | | Deposit date: | 2016-12-29 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural insights into a multifunctional inhibitor, 'AMTIN' from tubers of Alocasia macrorrhizos and its possible role in dengue protease (NS2B-NS3) inhibition.
Int. J. Biol. Macromol., 113, 2018
|
|
7Q93
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, NAD complex. | | Descriptor: | GLYCEROL, NADQ transcription factor, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
1QRP
 
 | | Human pepsin 3A in complex with a phosphonate inhibitor IVA-VAL-VAL-LEU(P)-(O)PHE-ALA-ALA-OME | | Descriptor: | PEPSIN 3A, methyl N-[(2S)-2-({(S)-hydroxy[(1R)-3-methyl-1-{[N-(3-methylbutanoyl)-L-valyl-L-valyl]amino}butyl]phosphoryl}oxy)-3-phenylpropanoyl]-L-alanyl-L-alaninate | | Authors: | Fujinaga, M, Cherney, M.M, Tarasova, N.I, Bartlett, P.A, Hanson, J.E, James, M.N.G. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QT1
 
 | |
