5YIP
 
 | | Crystal Structure of AnkG LIR/GABARAPL1 complex | | Descriptor: | Ankyrin-3, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
6Q2Y
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
5YOA
 
 | |
8SM0
 
 | | Crystal structure of human complement receptor 2 (CD21) in complex with Epstein-Barr virus major glycoprotein gp350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement receptor type 2, Envelope glycoprotein gp350, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
6QHM
 
 | | 14-3-3 sigma with RelA/p65 binding site pS281 | | Descriptor: | 14-3-3 protein sigma, LEU-SEP-GLU | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
6QK0
 
 | |
5YO5
 
 | |
100D
 
 | |
5YOE
 
 | | Crystal Structure of flavodoxin with engineered disulfide bond A43C-L74C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YIL
 
 | | Hoisting-loop in bacterial multidrug exporter AcrB is a highly flexible hinge that enables the large motion of the subdomains | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | Zwama, M, Sakurai, K, Hayashi, K, Nakashima, R, Kitagawa, K, Nishino, K, Yamaguchi, A. | | Deposit date: | 2017-10-05 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hoisting-Loop in Bacterial Multidrug Exporter AcrB Is a Highly Flexible Hinge That Enables the Large Motion of the Subdomains.
Front Microbiol, 8, 2017
|
|
6Q46
 
 | |
8SM1
 
 | | CRYSTAL STRUCTURE OF HUMAN ANTIBODY 769A9 IN COMPLEX WITH EPSTEIN-BARR VIRUS MAJOR GLYCOPROTEIN GP350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769A9 Fab heavy chain, 769A9 Fab light chain, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
8SH7
 
 | |
5YR1
 
 | | Crystal Structure of Cypovirus Polyhedra R13A/E73C/Y83C/S193C/A194C Mutant | | Descriptor: | 1,2-ETHANEDIOL, Polyhedrin | | Authors: | Negishi, H, Abe, S, Yamashita, K, Hirata, K, Niwase, K, Boudes, M, Coulibaly, F, Mori, H, Ueno, T. | | Deposit date: | 2017-11-08 | | Release date: | 2018-02-21 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Supramolecular protein cages constructed from a crystalline protein matrix
Chem. Commun. (Camb.), 54, 2018
|
|
6QJM
 
 | | Cryo-EM structure of heparin-induced 2N4R tau twister filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
6QJQ
 
 | | Cryo-EM structure of heparin-induced 2N3R tau filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
5YRL
 
 | | PPL3C-trehalose complex | | Descriptor: | PPL3-b, SULFATE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Nakae, S, Shionyu, M, Ogawa, T, Shirai, T. | | Deposit date: | 2017-11-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization
Proteins, 86, 2018
|
|
5YJS
 
 | | Structure of vicilin from Capsicum annuum | | Descriptor: | 2-HYDROXYBENZOIC ACID, CHLORIDE ION, COPPER (I) ION, ... | | Authors: | Shikhi, M, Nair, D.T, Salunke, D.M. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-guided identification of function: role ofCapsicum annuumvicilin during oxidative stress.
Biochem. J., 475, 2018
|
|
7X5C
 
 | | Solution structure of Tetrahymena p75OB1-p50PBM | | Descriptor: | Telomerase associated protein p50PBM, Telomerase-associated protein p75OB1 | | Authors: | Wu, B, Tang, T, Xue, H.J, Wu, J, Lei, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Association of the CST complex and p50 in Tetrahymena is crucial for telomere maintenance
Structure, 2022
|
|
6QGR
 
 | | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5YKR
 
 | |
5YSP
 
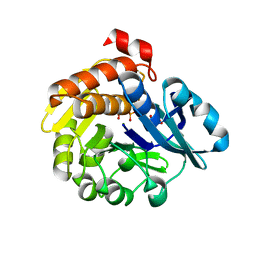 | | Pyrophosphate-dependent kinase in the ribokinase family complexed with a pyrophosphate analog and myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, MAGNESIUM ION, METHYLENEDIPHOSPHONIC ACID, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants.
Nat Commun, 9, 2018
|
|
8SM2
 
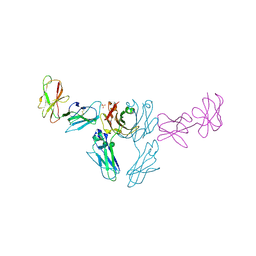 | |
6QK7
 
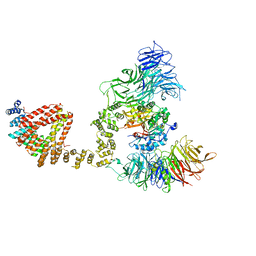 | | Elongator catalytic subcomplex Elp123 lobe | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Dauden, M.I, Jaciuk, M, Glatt, S. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of tRNA recognition by the Elongator complex.
Sci Adv, 5, 2019
|
|
7FLT
 
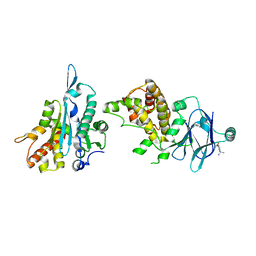 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G04 from the F2X-Universal Library | | Descriptor: | (1S)-1-(3-fluoro-4-methoxyphenyl)ethan-1-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
