1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
1EKF
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 1.95 ANGSTROMS (ORTHORHOMBIC FORM) | | Descriptor: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-08 | | Release date: | 2001-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EL2
 
 | |
8WG3
 
 | | mouse TMEM63b in LMNG-CHS micelle | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
1EVT
 
 | | CRYSTAL STRUCTURE OF FGF1 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 1 (FGFR1) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 1), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 1), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
1EWC
 
 | | CRYSTAL STRUCTURE OF ZN2+ LOADED STAPHYLOCOCCAL ENTEROTOXIN H | | Descriptor: | ENTEROTOXIN H, ZINC ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
6HLW
 
 | |
1EX9
 
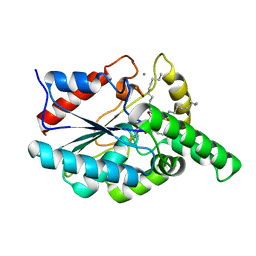 | | CRYSTAL STRUCTURE OF THE PSEUDOMONAS AERUGINOSA LIPASE COMPLEXED WITH RC-(RP,SP)-1,2-DIOCTYLCARBAMOYL-GLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, LACTONIZING LIPASE, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER | | Authors: | Nardini, M, Lang, D.A, Liebeton, K, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2000-05-02 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases.
J.Biol.Chem., 275, 2000
|
|
8WM5
 
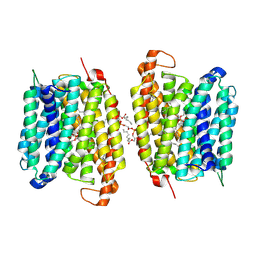 | |
1ESN
 
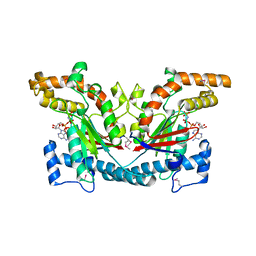 | | STRUCTURAL BASIS FOR THE FEEDBACK REGULATION OF ESCHERICHIA COLI PANTOTHENATE KINASE BY COENZYME A | | Descriptor: | MAGNESIUM ION, PANTOTHENATE KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, M, Park, C.G, Kim, J.Y, Rock, C.O, Jackowski, S, Park, H.W. | | Deposit date: | 2000-04-10 | | Release date: | 2000-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A.
J.Biol.Chem., 275, 2000
|
|
1EV2
 
 | | CRYSTAL STRUCTURE OF FGF2 IN COMPLEX WITH THE EXTRACELLULAR LIGAND BINDING DOMAIN OF FGF RECEPTOR 2 (FGFR2) | | Descriptor: | PROTEIN (FIBROBLAST GROWTH FACTOR 2), PROTEIN (FIBROBLAST GROWTH FACTOR RECEPTOR 2), SULFATE ION | | Authors: | Plotnikov, A.N, Hubbard, S.R, Schlessinger, J, Mohammadi, M. | | Deposit date: | 2000-04-19 | | Release date: | 2000-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity.
Cell(Cambridge,Mass.), 101, 2000
|
|
8WG4
 
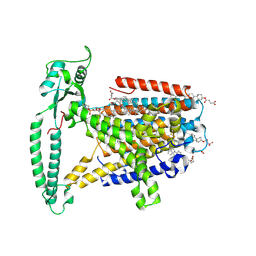 | | mouse TMEM63b in DDM-CHS micelle with YN9303-24 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
1EXB
 
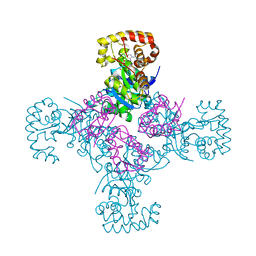 | | STRUCTURE OF THE CYTOPLASMIC BETA SUBUNIT-T1 ASSEMBLY OF VOLTAGE-DEPENDENT K CHANNELS | | Descriptor: | KV BETA2 PROTEIN, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM CHANNEL KV1.1 | | Authors: | Gulbis, J.M, Zhou, M, Mann, S, MacKinnon, R. | | Deposit date: | 2000-05-02 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels.
Science, 289, 2000
|
|
8WFM
 
 | |
6HSG
 
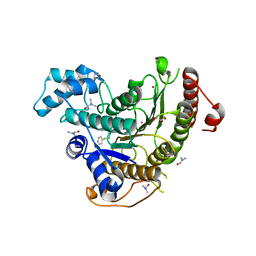 | | Crystal structure of Schistosoma mansoni HDAC8 H292M mutant complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
8WD7
 
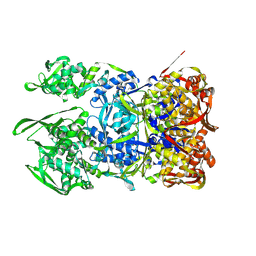 | | Structural organization of the palisade layer in isolated vaccinia virus cores | | Descriptor: | Core protein OPG136 | | Authors: | Liu, Y, Qu, X, Duan, M, Shi, X, Liu, S, Shi, Y, Gao, G.F. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Cryo-ET reveals A10 protein as a major component of the poxvirus palisade layer
To Be Published
|
|
8WDQ
 
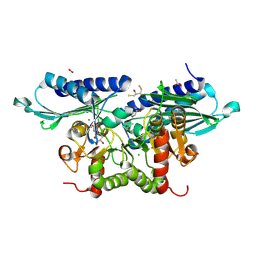 | | Crystal Structure of Pseudomonas aeruginosa SuhB in complex with D-myo-inositol-1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadav, V.K, Maji, S, Shukla, M, Bhattacharyya, S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Pseudomonas aeruginosa SuhB in complex with D-myo-inositol-1-phosphate
To Be Published
|
|
7UE1
 
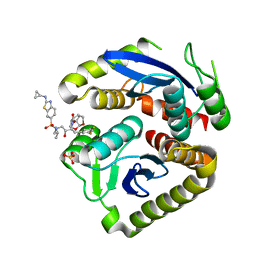 | | HIV-1 Integrase Catalytic Core Domain Mutant (KGD) in Complex with Inhibitor GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Integrase, SULFATE ION | | Authors: | Aoki, M, Aoki-Ogata, H, Bulut, H, Hayashi, H, Davis, D, Hasegawa, K, Yarchoan, R, Ghosh, A.K, Pau, A.K, Mitsuya, H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants.
Sci Adv, 9, 2023
|
|
6HT6
 
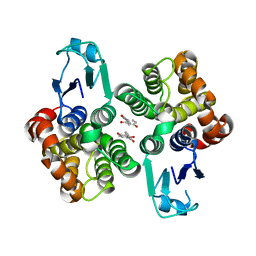 | | Crystal structure of glutathione transferase Omega 2S from Trametes versicolor in complex with 2,4-dihydroxybenzophenone | | Descriptor: | Glutathione S-transferase omega 2S, MAGNESIUM ION, [2,4-bis(oxidanyl)phenyl]-phenyl-methanone | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
6HU1
 
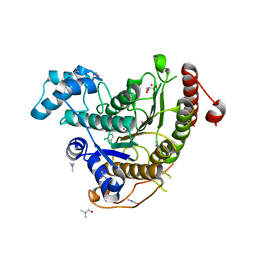 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 10 | | Descriptor: | 4-chloranyl-3-[(2,4-dichlorophenyl)carbonylamino]-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
7UES
 
 | | PANK3 complex structure with compound PZ-4202 | | Descriptor: | MAGNESIUM ION, N-(4-{2-[4-(6-cyanopyridazin-3-yl)piperazin-1-yl]-2-oxoethyl}phenyl)methanesulfonamide, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of hPANK Activators with Improved Pharmacological Properties
To Be Published
|
|
8XAC
 
 | |
1EYX
 
 | | CRYSTAL STRUCTURE OF R-PHYCOERYTHRIN AT 2.2 ANGSTROMS | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOUROBILIN, ... | | Authors: | Contreras-Martel, C, Legrand, P, Piras, C, Vernede, X, Martinez-Oyanedel, J, Bunster, M, Fontecilla-Camps, J.C. | | Deposit date: | 2000-05-09 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallization and 2.2 A resolution structure of R-phycoerythrin from Gracilaria chilensis: a case of perfect hemihedral twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7UER
 
 | |
1EGD
 
 | | STRUCTURE OF T255E, E376G MUTANT OF HUMAN MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MEDIUM CHAIN ACYL-COA DEHYDROGENASE | | Authors: | Lee, H.J, Wang, M, Paschke, R, Nandy, A, Ghisla, S, Kim, J.P. | | Deposit date: | 1996-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity.
Biochemistry, 35, 1996
|
|
