1EET
 
 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
8WT6
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
7UNG
 
 | | 48-nm repeat of the human respiratory doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 161, Cilia- and flagella-associated protein 20, Cilia- and flagella-associated protein 45, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-11 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1EKV
 
 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL): THREE DIMENSIONAL STRUCTURE OF ENZYME INACTIVATED BY TRIS BOUND TO THE PYRIDOXAL-5'-PHOSPHATE ON ONE END AND ACTIVE SITE LYS202 NZ ON THE OTHER. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7UN1
 
 | | 8-nm repeat of the human sperm tip singlet microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gui, M, Croft, J.T, Zabeo, D, Acharya, V, Kollman, J.M, Burgoyne, T, Hoog, J.L, Brown, A. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1ELN
 
 | |
1EKG
 
 | | MATURE HUMAN FRATAXIN | | Descriptor: | FRATAXIN | | Authors: | Dhe-Paganon, S, Shigeta, R, Chi, Y.I, Ristow, M, Shoelson, S.E. | | Deposit date: | 2000-03-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human frataxin.
J.Biol.Chem., 275, 2000
|
|
1EKP
 
 | | CRYSTAL STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 2.5 ANGSTROMS (MONOCLINIC FORM). | | Descriptor: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7PAG
 
 | | The pore conformation of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Perforin-1 | | Authors: | Ivanova, M.E, Lukoyanova, N, Malhotra, S, Topf, M, Trapani, J.A, Voskoboinik, I, Saibil, H.R. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The pore conformation of lymphocyte perforin.
Sci Adv, 8, 2022
|
|
8WT9
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution) | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
1EAO
 
 | | THE RUNX1 Runt domain at 1.4A resolution: a structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | BROMIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T.H, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-14 | | Release date: | 2002-09-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Runx1 Runt Domain at 1.25A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
8UA2
 
 | | Crystal Structure of infected cell protein 0 (ICP0) from herpes simplex virus 1 (proteolyzed fragment) | | Descriptor: | IODIDE ION, RL2 | | Authors: | Lovell, S, Kashipathy, M, Battaile, K.P, Cooper, A, Davido, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
Proteins, 92, 2024
|
|
7UT3
 
 | | Crystal structure of complex of Fab, G10C with GalNAc-pNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-nitrophenyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside, Fab protein heavy chain, ... | | Authors: | Li, M, Wlodawer, A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Development of a GalNAc-Tyrosine-Specific Monoclonal Antibody and Detection of Tyrosine O -GalNAcylation in Numerous Human Tissues and Cell Lines.
J.Am.Chem.Soc., 144, 2022
|
|
1EP9
 
 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CONFORMATIONAL CHANGE | | Descriptor: | ORNITHINE TRANSCARBAMYLASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Morizono, H, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2000-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes.
Biochem.J., 354, 2001
|
|
4THN
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-THROMBIN-HIRUNORM IV COMPLEX REVEALS A NOVEL SPECIFICITY SITE RECOGNITION MODE. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUNORM IV | | Authors: | Lombardi, A, De Simone, G, Nastri, F, Galdiero, S, Della Morte, R, Staiano, N, Pedone, C, Bolognesi, M, Pavone, V. | | Deposit date: | 1998-09-18 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of alpha-thrombin-hirunorm IV complex reveals a novel specificity site recognition mode.
Protein Sci., 8, 1999
|
|
7Q0L
 
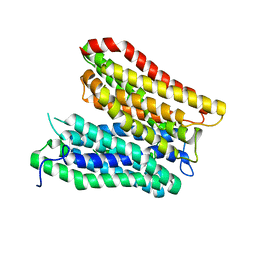 | | Crystal structure of the peptide transporter YePEPT-K314A at 2.93 A | | Descriptor: | Peptide transporter YePEPT | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Boggavarapu, R, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
7Q0M
 
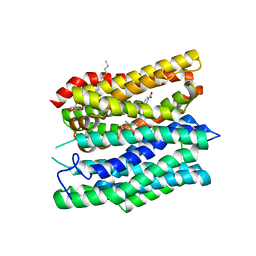 | | Crystal structure of the peptide transporter YePEPT-K314A in complex with LZNV at 2.66 A | | Descriptor: | (2~{S})-2-[[(2~{S})-2-azanyl-6-[(4-nitrophenyl)methoxycarbonylamino]hexanoyl]amino]-3-methyl-butanoic acid, Peptide transporter YePEPT, UNDECYL-MALTOSIDE | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
7PKR
 
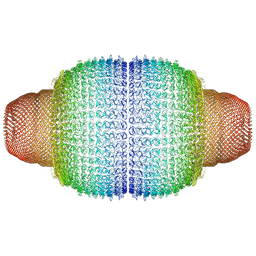 | | Vault structure in primmed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PKZ
 
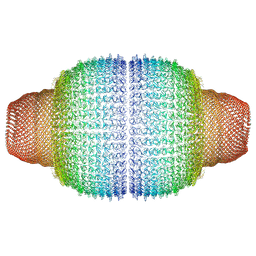 | | Vault structure in committed conformation | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7PO6
 
 | | Xist (m6A)UCG tetraloop RNA bound to the YTH domain of YTHDC1 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-TRIPHOSPHATE, Isoform 2 of YTH domain-containing protein 1, ... | | Authors: | Jones, A.N, Mourao, A, Sattler, M. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural effects of m6A modification of the Xist A-repeat AUCG tetraloop and its recognition by YTHDC1.
Nucleic Acids Res., 50, 2022
|
|
7PUJ
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, ZINC ION | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
6H67
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HUV
 
 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 39 | | Descriptor: | (2~{S})-~{N}-[(3~{S},4~{R})-1-cyclohexyl-5-methyl-4,5-bis(oxidanyl)hexan-3-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
7PKY
 
 | | Half-vault structure | | Descriptor: | Major vault protein | | Authors: | Guerra, P, Gonzalez-Alamos, M, Llauro, A, Casanas, A, Querol-Audi, J, de Pablo, P, Verdaguer, N. | | Deposit date: | 2021-08-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Symmetry disruption commits vault particles to disassembly.
Sci Adv, 8, 2022
|
|
7UWS
 
 | | Atomic model of the partial VSV nucleocapsid | | Descriptor: | Matrix protein, Nucleoprotein, RNA | | Authors: | Zhou, K, Si, Z, Ge, P, Tsao, J, Luo, M, Zhou, Z.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Atomic model of vesicular stomatitis virus and mechanism of assembly.
Nat Commun, 13, 2022
|
|
