6ZDV
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 5d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6QMI
 
 | |
5Z2I
 
 | | Structure of Dictyostelium discoideum mitochondrial calcium uniporter N-ternimal domain (Se-DdMCU-NTD) | | Descriptor: | Dictyostelium discoideum mitochondrial calcium uniporter | | Authors: | Yuan, Y, Wen, M, Chou, J, Li, D, Bo, O. | | Deposit date: | 2018-01-02 | | Release date: | 2019-01-02 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Characterization of the N-Terminal Domain of theDictyostelium discoideumMitochondrial Calcium Uniporter.
Acs Omega, 5, 2020
|
|
5HMB
 
 | | Crystal structure of S. sahachiroi AziG | | Descriptor: | Azi13, SULFATE ION | | Authors: | Erb, M.S, Zhang, Y, Ealick, S.E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Polyketide Ring Expansion Mediated by a Thioesterase, Chain Elongation and Cyclization Domain, in Azinomycin Biosynthesis: Characterization of AziB and AziG.
Biochemistry, 55, 2016
|
|
4MY8
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Gorla, S.K, Kavitha, M, Cuny, G, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2924 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor Q21
To be Published, 2013
|
|
8QUC
 
 | | Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1 | | Descriptor: | (5R)-5-ethyl-3-[6-(3-methoxy-4-methyl-phenoxy)pyridin-3-yl]imidazolidine-2,4-dione, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, G, Mckinley, G, Marsden, B, Pike, A.C.W, Ye, M, Brooke, L.M, Bakshi, S, Pilati, N, Marasco, A, Gunthorpe, M, Alvaro, G, Large, C, Lakshminaraya, B, Williams, E, Sauer, D.B. | | Deposit date: | 2023-10-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The binding and mechanism of a positive allosteric modulator of Kv3 channels.
Nat Commun, 15, 2024
|
|
5ZH5
 
 | | CRYSTAL STRUCTURE OF PfKRS WITH INHIBITOR CLADO-2 | | Descriptor: | (3S)-6,8-dihydroxy-3-{[(2R,6S)-6-methyloxan-2-yl]methyl}-3,4-dihydro-1H-2-benzopyran-1-one, CHLORIDE ION, LYSINE, ... | | Authors: | Babbar, P, Malhotra, N, Sharma, M, Harlos, K, Reddy, D.S, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific Stereoisomeric Conformations Determine the Drug Potency of Cladosporin Scaffold against Malarial Parasite
J. Med. Chem., 61, 2018
|
|
3DHM
 
 | | Beta 2 microglobulin mutant D59P | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Colombo, M, de Rosa, M, Bolognesi, M, Giorgetti, S, Bellotti, V. | | Deposit date: | 2008-06-18 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DE loop mutations affect beta2-microglobulin stability and amyloid aggregation
Biochem.Biophys.Res.Commun., 377, 2008
|
|
4N49
 
 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
5ZN1
 
 | | X-ray structure of protein kinase ck2 alpha subunit in D2O | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN5
 
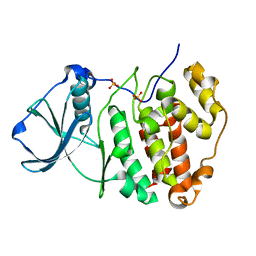 | | X-ray structure of protein kinase ck2 alpha subunit H148A mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
8QQU
 
 | |
5ZOK
 
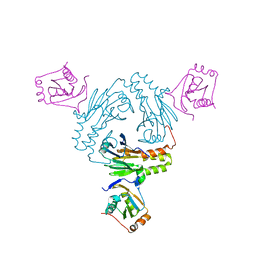 | | Crystal structure of human SMAD1-MAN1 complex. | | Descriptor: | Inner nuclear membrane protein Man1, Mothers against decapentaplegic homolog 1 | | Authors: | Miyazono, K, Ito, T, Tanokura, M. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for receptor-regulated SMAD recognition by MAN1
Nucleic Acids Res., 46, 2018
|
|
8QAW
 
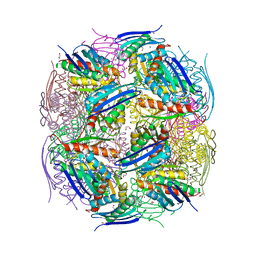 | | Medicago truncatula HISN5 (IGPD) in complex with MN, IMD, EDO, FMT, GOL and TRS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAX
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, GOL and TRS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
6BOW
 
 | |
1A11
 
 | | NMR STRUCTURE OF MEMBRANE SPANNING SEGMENT 2 OF THE ACETYLCHOLINE RECEPTOR IN DPC MICELLES, 10 STRUCTURES | | Descriptor: | ACETYLCHOLINE RECEPTOR M2 | | Authors: | Gesell, J.J, Sun, W, Montal, M, Opella, S.J. | | Deposit date: | 1997-12-19 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
8QG0
 
 | | Archaeoglobus fulgidus AfAgo complex with AfAgo-N protein (fAfAgo) bound with 17 nt RNA guide and 17 nt DNA target | | Descriptor: | AfAgo-N protein, DNA target 17 nt, Piwi protein, ... | | Authors: | Manakova, E.N, Zaremba, M, Pocevicuite, R, Golovinas, E, Zagorskaite, E, Silanskas, A. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
8R5Q
 
 | | Structure of apo TDO with a bound inhibitor | | Descriptor: | 3-chloranyl-~{N}-[(1~{S})-1-(6-chloranylpyridin-3-yl)-2-phenyl-ethyl]aniline, Tryptophan 2,3-dioxygenase, alpha-methyl-L-tryptophan | | Authors: | Wicki, M, Mac Sweeney, A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery and binding mode of a small molecule inhibitor of the apo form of human TDO2
Biorxiv, 2024
|
|
6YT5
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15-gp16 core complex intermediate assembly | | Descriptor: | Internal virion protein gp15, Peptidoglycan transglycosylase gp16 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YSZ
 
 | | Cryo-EM structure of T7 bacteriophage DNA translocation gp15 core protein intermediate assembly | | Descriptor: | Internal virion protein gp15 | | Authors: | Perez-Ruiz, M, Pulido-Cid, M, Luque-Ortega, J.R, Cuervo, A, Carrascosa, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5Z3N
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base 5fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(5FC)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2018-01-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5Z3W
 
 | | Malate dehydrogenase binds silver at C113 | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm
Chem Sci, 2020
|
|
3AZ6
 
 | | Crystal structure of Co/T-HEWL | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Abe, S, Tsujimoto, M, Yoneda, K, Ohba, M, Hikage, T, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2011-05-20 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Porous protein crystals as reaction vessels for controlling magnetic properties of nanoparticles
Small, 8, 2012
|
|
6OWN
 
 | |
