7ZMT
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
6YDB
 
 | | Human wtSTING in complex with 2',2'-difluoro-3',3'-c-di-GMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-17-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ZMV
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
6YDZ
 
 | | Human wtSTING in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ZMP
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-055 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-055, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.626 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMR
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-011 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-011, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
8TTW
 
 | | Cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tolbert, W.D, Pozharski, E, Pazgier, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
6YEA
 
 | | Human wtSTING in complex with 2',2'-difluoro-3',3'-cGAMP | | Descriptor: | 2',2'-difluoro-3',3'-cGAMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7ZMQ
 
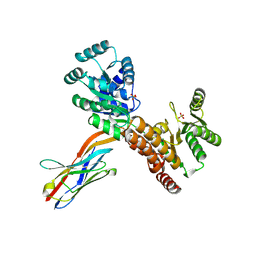 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZML
 
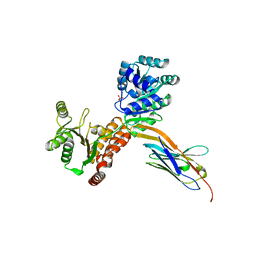 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G1-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G1-001, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMM
 
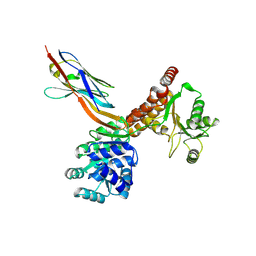 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2-001, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMN
 
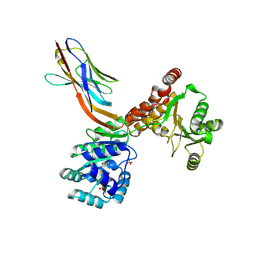 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-048 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-048, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7QIO
 
 | | Homology model of myosin neck domain in skeletal sarcomere | | Descriptor: | Myosin light chain 1/3, skeletal muscle isoform, Myosin regulatory light chain 2, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
2VF3
 
 | | Aquifex aeolicus IspE in complex with ligand | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of the Kinase Ispe: Structure-Activity Relationships and Co-Crystal Structure Analysis.
Org.Biomol.Chem., 6, 2008
|
|
3E1Z
 
 | | Crystal structure of the parasite protesase inhibitor chagasin in complex with papain | | Descriptor: | ACETIC ACID, Chagasin, FORMIC ACID, ... | | Authors: | Redzynia, I, Bujacz, G, Bujacz, A, Ljunggren, A, Abrahamson, M, Jaskolski, M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the parasite inhibitor chagasin in complex with papain allows identification of structural requirements for broad reactivity and specificity determinants for target proteases.
Febs J., 276, 2009
|
|
3DL3
 
 | | Crystal structure of the tellurite resistance protein TehB. Northeast Structural Genomics Consortium target VfR98 . | | Descriptor: | Tellurite resistance protein B | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the tellurite resistance protein TehB. Northeast Structural Genomics Consortium target VfR98.
To be Published
|
|
7QIN
 
 | | In situ structure of actomyosin complex in skeletal sarcomere | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
3DIB
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (I106A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DM3
 
 | | Crystal structure of a domain of a Replication factor A protein, from Methanocaldococcus jannaschii. NorthEast Structural Genomics target MjR118E | | Descriptor: | Replication factor A, SODIUM ION | | Authors: | Seetharaman, J, Su, M, Maglaqui, M, Janjua, H, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a domain of a Replication factor A protein, from Methanocaldococcus jannaschii. NorthEast Structural Genomics target MjR118E
To be Published
|
|
5KSU
 
 | | Crystal structure of HLA-DQ2.5-CLIP1 at 2.73 resolution | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DQ alpha 1 chain, ... | | Authors: | Nguyen, T.-B, Jayaraman, P, Bergseng, E, Madhusudhan, M.S, Kim, C.-Y, Sollid, L.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Unraveling the structural basis for the unusually rich association of human leukocyte antigen DQ2.5 with class-II-associated invariant chain peptides.
J. Biol. Chem., 292, 2017
|
|
5KUG
 
 | | Human mitochondrial calcium uniporter (residues 72-189) crystal structure with lithium | | Descriptor: | Calcium uniporter protein, mitochondrial | | Authors: | Mok, M.C.Y, Lee, S.K, Junop, M.S, Stathopulos, P.B. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations.
Cell Chem Biol, 23, 2016
|
|
6Y0D
 
 | |
6HB4
 
 | | TFAM in Complex with Site-Y | | Descriptor: | DNA (5'*CP*TP*GP*TP*GP*CP*AP*GP*AP*CP*AP*TP*TP*CP*AP*AP*TP*TP*GP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*AP*AP*TP*TP*GP*AP*AP*TP*GP*TP*CP*TP*GP*CP*AP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Cuppari, A, Fernandez-Millan, P, Rubio-Cosials, A, Tarres-Sole, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
6YQ5
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
7PAY
 
 | | Structure of the human heterotetrameric cis-prenyltransferase complex in complex with magnesium and GGsPP | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit DHDDS, Dehydrodolichyl diphosphate synthase complex subunit NUS1, MAGNESIUM ION, ... | | Authors: | Giladi, M, Lisnyansky Bar-El, M, Haitin, Y. | | Deposit date: | 2021-07-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for long-chain isoprenoid synthesis by cis -prenyltransferases.
Sci Adv, 8, 2022
|
|
