2VA5
 
 | | X-ray crystal structure of beta secretase complexed with compound 8c | | Descriptor: | 2-amino-6-[2-(1H-indol-6-yl)ethyl]pyrimidin-4(3H)-one, BETA-SECRETASE 1 ., IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
4EC4
 
 | | XIAP-BIR3 in complex with a potent divalent Smac mimetic | | Descriptor: | (3S,6S,7S,9aS,3'S,6'S,7'S,9a'S)-N,N'-(benzene-1,4-diylbis{butane-4,1-diyl-1H-1,2,3-triazole-1,4-diyl[(S)-phenylmethanediyl]})bis[7-(hydroxymethyl)-6-{[(2S)-2-(methylamino)butanoyl]amino}-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide], 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Baculoviral IAP repeat-containing protein 4, ... | | Authors: | Mastrangelo, E, Cossu, F, Bolognesi, M, Milani, M. | | Deposit date: | 2012-03-26 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insight into inhibitor of apoptosis proteins recognition by a potent divalent smac-mimetic.
Plos One, 7, 2012
|
|
4C2U
 
 | | Crystal structure of Deinococcus radiodurans UvrD in complex with DNA, Form 1 | | Descriptor: | DNA HELICASE II, FOR25, GLYCEROL, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
4EE9
 
 | | Crystal structure of the RBcel1 endo-1,4-glucanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase | | Authors: | Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-03 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Three-dimensional structure of RBcel1, a metagenome-derived psychrotolerant family GH5 endoglucanase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2MI6
 
 | |
4CCY
 
 | | Crystal structure of carboxylesterase CesB (YbfK) from Bacillus subtilis | | Descriptor: | CARBOXYLESTERASE YBFK, SODIUM ION | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
4EFN
 
 | | Crystal structure of H-Ras Q61L in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
2MND
 
 | | Recognition complex of DNA d(CGACGCGTCG)2 with thiazotropsin B | | Descriptor: | 5'-D(*CP*GP*AP*CP*GP*CP*GP*TP*CP*G)-3', thiazotropsin B | | Authors: | Alniss, H.Y, Salvia, M.-V, Sadikov, M, Golovchenko, I, Anthony, N.G, Khalaf, A.I, Mackay, S.P, Suckling, C.J, Parkinson, J.A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the DNA minor groove by thiazotropsin analogues.
Chembiochem, 15, 2014
|
|
2MNX
 
 | | Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA adduct induced by 1,2-epoxy-3-butene | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(6HB)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Kotapati, S, Turo, M, Tretyakova, N, Stone, M.P. | | Deposit date: | 2014-04-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene.
Chem.Res.Toxicol., 27, 2014
|
|
2M1V
 
 | |
4BTT
 
 | | factor Xa in complex with the dual thrombin-FXa inhibitor 31. | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, COAGULATION FACTOR X LIGHT CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
4C2T
 
 | | Crystal structure of full length Deinococcus radiodurans UvrD in complex with DNA | | Descriptor: | DNA HELICASE II, DNA STRAND FOR28, DNA STRAND REV28, ... | | Authors: | Stelter, M, Acajjaoui, S, McSweeney, S, Timmins, J. | | Deposit date: | 2013-08-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.997 Å) | | Cite: | Structural and Mechanistic Insight Into DNA Unwinding by Deinococcus Radiodurans Uvrd.
Plos One, 8, 2013
|
|
2MNB
 
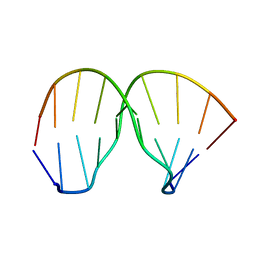 | | Thiazotropsin B DNA recognition sequence d(CGACGCGTCG)2 | | Descriptor: | 5'-D(*CP*GP*AP*CP*GP*CP*GP*TP*CP*G)-3' | | Authors: | Alniss, H.Y, Salvia, M.-V, Sadikov, M, Golovchenko, I, Anthony, N.G, Khalaf, A.I, Mackay, S.P, Suckling, C.J, Parkinson, J.A. | | Deposit date: | 2014-04-02 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the DNA minor groove by thiazotropsin analogues.
Chembiochem, 15, 2014
|
|
2MNE
 
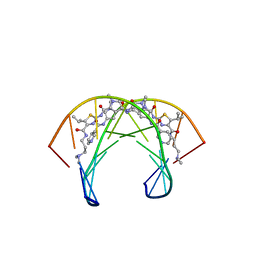 | | Recognition complex of DNA d(CGACTAGTCG)2 with thiazotropsin analogue AIK-18/51 | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3', AIK-18/51 | | Authors: | Alniss, H.Y, Salvia, M.-V, Sadikov, M, Golovchenko, I, Anthony, N.G, Khalaf, A.I, Mackay, S.P, Suckling, C.J, Parkinson, J.A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the DNA minor groove by thiazotropsin analogues.
Chembiochem, 15, 2014
|
|
4C8Y
 
 | |
4BTU
 
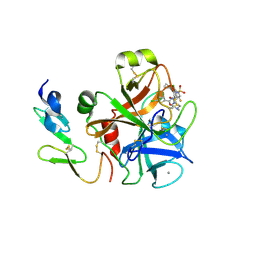 | | Factor Xa in complex with the dual thrombin-FXa inhibitor 57. | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid [(S)-2-[2-chloro-5-fluoro-3-(2-oxo-piperidin-1-yl)-benzenesulfonylamino]-3-(4-methyl-piperazin-1-yl)-3-oxo-propyl]-amide, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Meneyrol, J, Follmann, M, Lassalle, G, Wehner, V, Barre, G, Rousseaux, T, Altenburger, J.M, Petit, F, Bocskei, Z, Stehlin-Gaon, C, Schreuder, H, Alet, N, Herault, J.-P, Millet, L, Dol, F, Hasbrand, C, Schaeffer, P, Sadoun, F, Klieber, S, Briot, C, Bono, F, Herbert, J.-M. | | Deposit date: | 2013-06-19 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 5-Chlorothiophene-2-Carboxylic Acid [(S)-2-[2-Methyl-3-(2-Oxopyrrolidin-1-Yl)Benzenesulfonylamino]-3-(4-Methylpiperazin-1-Yl)-3-Oxopropyl]Amide (Sar107375), a Selective and Potent Orally Active Dual Thrombin and Factor Xa Inhibitor.
J.Med.Chem., 56, 2013
|
|
4BS5
 
 | | MOUSE CATHEPSIN S WITH COVALENT LIGAND | | Descriptor: | (2S,4R)-N-[1-(iminomethyl)cyclopropyl]-4-[2-(trifluoromethyl)phenyl]sulfonyl-pyrrolidine-2-carboxamide, CATHEPSIN S | | Authors: | Banner, D.W, Benz, J, Gsell, B, Stihle, M, Ruf, A, Haap, W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Identification of Potent and Selective Cathepsin S (Cats) Inhibitors Containing Different Central Cyclic Scaffolds.
J.Med.Chem., 56, 2013
|
|
4EA6
 
 | | Crystal structure of Fungal lipase from Thermomyces(Humicola) lanuginosa at 2.30 Angstrom resolution. | | Descriptor: | Lipase | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Fungal lipase from Thermomyces(Humicola) lanuginosa at 2.30 Angstrom resolution.
To be Published
|
|
4BQG
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-(3,4-dichloro-phenoxy)-benzene-1,3-diol, HSP90AA1 PROTEIN | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
4BUE
 
 | | Crystal structure of human tankyrase 2 in complex with 3-methyl-N-(4-(4-oxo-3,4-dihydroquinazolin-2-yl)phenyl)butanamide | | Descriptor: | 3-METHYL-N-[4-(4-OXO-3,4-DIHYDROQUINAZOLIN-2-YL)PHENYL)BUTANAMIDE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
2M2D
 
 | | Human programmed cell death 1 receptor | | Descriptor: | Programmed cell death protein 1 | | Authors: | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-27 | | Last modified: | 2013-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
2ML5
 
 | |
2M3L
 
 | |
4C1N
 
 | | Corrinoid protein reactivation complex with activator | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE CORRINOID/IRON-SULFUR PROTEIN, GAMMA SUBUNIT, CO DEHYDROGENASE/ACETYL-COA SYNTHASE, ... | | Authors: | Hennig, S.E, Goetzl, S, Jeoung, J.H, Bommer, M, Lendzian, F, Hildebrandt, P, Dobbek, H. | | Deposit date: | 2013-08-13 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | ATP-Induced Electron Transfer by Redox-Selective Partner Recognition
Nat.Commun., 5, 2014
|
|
4BWK
 
 | | Structure of Neurospora crassa PAN3 pseudokinase | | Descriptor: | PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN-3, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Christie, M, Boland, A, Huntzinger, E, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2013-07-04 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Pan3 Pseudokinase Reveals the Basis for Interactions with the Pan2 Deadenylase and the Gw182 Proteins
Mol.Cell, 51, 2013
|
|
