2XYS
 
 | | Crystal structure of Aplysia californica AChBP in complex with strychnine | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR, STRYCHNINE | | Authors: | Brams, M, Pandya, A, Kuzmin, D, van Elk, R, Krijnen, L, Yakel, J.L, Tsetlin, V, Smit, A.B, Ulens, C. | | Deposit date: | 2010-11-19 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | A Structural and Mutagenic Blueprint for Molecular Recognition of Strychnine and D-Tubocurarine by Different Cys-Loop Receptors.
Plos Biol., 9, 2011
|
|
3NJG
 
 | | K98A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
246D
 
 | | STRUCTURE OF THE PURINE-PYRIMIDINE ALTERNATING RNA DOUBLE HELIX, R(GUAUAUA)D(C) , WITH A 3'-TERMINAL DEOXY RESIDUE | | Descriptor: | DNA/RNA (5'-R(*GP*UP*AP*UP*AP*UP*AP*)-D(*C)-3'), SODIUM ION | | Authors: | Wahl, M.C, Ban, C, Sekharudu, C, Ramakrishnan, B, Sundaralingam, M. | | Deposit date: | 1996-01-25 | | Release date: | 1996-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the purine-pyrimidine alternating RNA double helix, r(GUAUAUA)d(C), with a 3'-terminal deoxy residue.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2XSV
 
 | | Crystal structure of L69A mutant Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
290D
 
 | |
3NJK
 
 | | D116A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis, at pH5.5 | | Descriptor: | GLYCEROL, Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
2Y0M
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MOF | | Descriptor: | ACETYL COENZYME *A, MALE-SPECIFIC LETHAL 1 HOMOLOG, PROBABLE HISTONE ACETYLTRANSFERASE MYST1, ... | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4I6K
 
 | | Crystal structure of probable 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE ABAYE1769 (TARGET EFI-505029) from Acinetobacter baumannii with citric acid bound | | Descriptor: | Amidohydrolase family protein, CITRIC ACID | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE (TARGET EFI-505029) FROM Acinetobacter baumannii
To be Published
|
|
2A08
 
 | | Structure of the yeast YHH6 SH3 domain | | Descriptor: | Hypothetical 41.8 kDa protein in SPO13-ARG4 intergenic region | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Lehmann, F, Zou, P, Wilmanns, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 3-D proteome of yeast SH3 domains
To be Published
|
|
2A0P
 
 | |
2Y0N
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN DOSAGE COMPENSATION FACTORS MSL1 AND MSL3 | | Descriptor: | MALE-SPECIFIC LETHAL 1 HOMOLOG, MALE-SPECIFIC LETHAL 3 HOMOLOG | | Authors: | Kadlec, J, Hallacli, E, Lipp, M, Holz, H, Sanchez Weatherby, J, Cusack, S, Akhtar, A. | | Deposit date: | 2010-12-04 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Mof and Msl3 Recruitment Into the Dosage Compensation Complex by Msl1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2Y3X
 
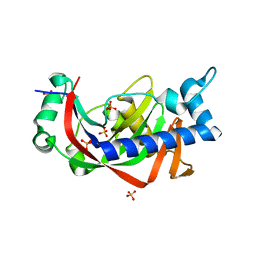 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, complexed with sulfate | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, GLYCEROL, ... | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2011-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Myelin 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase: Active-Site Ligand Binding and Molecular Conformation.
Plos One, 7, 2012
|
|
2A37
 
 | | Solution structure of the T22G mutant of N-terminal SH3 domain of DRK (DRKN SH3 DOMAIN) | | Descriptor: | Protein E(sev)2B | | Authors: | Bezsonova, I, Singer, A, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
4I54
 
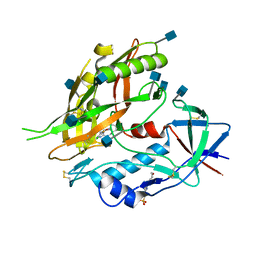 | |
4BGL
 
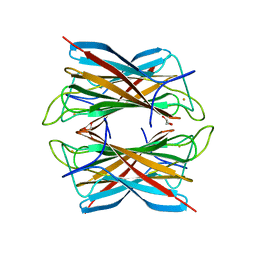 | | Superoxide reductase (Neelaredoxin) from Archaeoglobus fulgidus | | Descriptor: | FE (III) ION, GLYCEROL, SUPEROXIDE REDUCTASE | | Authors: | Bandeiras, T.M, Rodrigues, J.V, Sousa, C.M, Barradas, A.R, Pinho, F.G, Pinto, A.F, Teixeira, M, Matias, P.M, Romao, C.V. | | Deposit date: | 2013-03-27 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Superoxide Reductase from Giardia Intestinalis: Structural Characterization of the First Sor from a Eukaryotic Organism Shows an Iron Centre that is Highly Sensitive to Photoreduction
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4I9Q
 
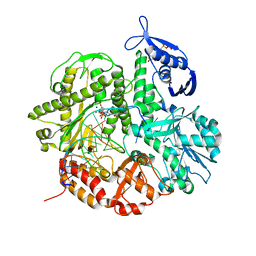 | | Crystal structure of the ternary complex of the D714A mutant of RB69 DNA polymerase | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3'), ... | | Authors: | Guja, K.E, Jacewicz, A, Trzemecka, A, Plochocka, D, Yakubovskaya, E, Bebenek, A, Garcia-Diaz, M. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Remote Palm Domain Residue of RB69 DNA Polymerase Is Critical for Enzyme Activity and Influences the Conformation of the Active Site.
Plos One, 8, 2013
|
|
4HPA
 
 | |
3NON
 
 | | Crystal Structure of Isocyanide Hydratase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, Isocyanide hydratase | | Authors: | Lakshminarasimhan, M, Madzelan, P, Nan, R, Milkovic, N.M, Wilson, M.A. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Evolution of New Enzymatic Function by Structural Modulation of Cysteine Reactivity in Pseudomonas fluorescens Isocyanide Hydratase.
J.Biol.Chem., 285, 2010
|
|
2A3V
 
 | | Structural basis for broad DNA-specificity in integron recombination | | Descriptor: | DNA (31-MER), DNA (34-MER), site-specific recombinase IntI4 | | Authors: | MacDonald, D, Demarre, G, Bouvier, M, Mazel, D, Gopaul, D.N. | | Deposit date: | 2005-06-27 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for broad DNA-specificity in integron recombination.
Nature, 440, 2006
|
|
2A50
 
 | | fluorescent protein asFP595, wt, off-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2XKL
 
 | | Crystal Structure of Mouse Apolipoprotein M | | Descriptor: | 1,2-ETHANEDIOL, APOLIPOPROTEIN M, GLYCEROL, ... | | Authors: | Sevvana, M, Kassler, K, Josefin, A, Weiler, S, Dahlback, B, Sticht, H, Muller, Y.A. | | Deposit date: | 2010-07-09 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mouse Apom Displays an Unprecedented Seven-Stranded Lipocalin Fold: Folding Decoy or Alternative Native Fold?
J.Mol.Biol., 404, 2010
|
|
2A6O
 
 | | Crystal Structure of the ISHp608 Transposase in Complex with Stem-loop DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*TP*AP*GP*CP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*GP*A)-3', ISHp608 Transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
4I7O
 
 | |
2XS8
 
 | | Crystal Structure of ALIX in complex with the SIVagmTan-1 AYDPARKLL Late Domain | | Descriptor: | PROGRAMMED CELL DEATH 6-INTERACTING PROTEIN, SIVAGMTAN-1 GAG P6 | | Authors: | Zhai, Q, Landesman, M, Robinson, H, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Identification and Structural Characterization of the Alix-Binding Late Domains of Sivmac239 and Sivagmtan-1.
J.Virol., 85, 2011
|
|
2A4W
 
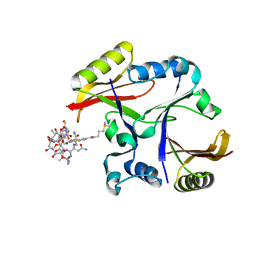 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Copper(II)-Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, COPPER (II) ION, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
