3SNN
 
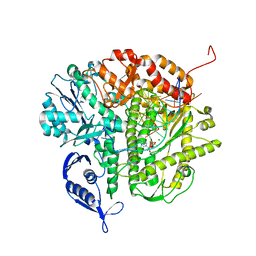 | | RB69 DNA Polymerase (L561A/S565G/Y567A) Ternary Complex with dCTP Opposite dG in the presence of Mg2+ | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3', 5'-D(*TP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
4KSO
 
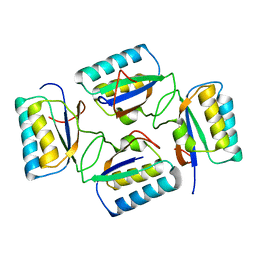 | |
3F4S
 
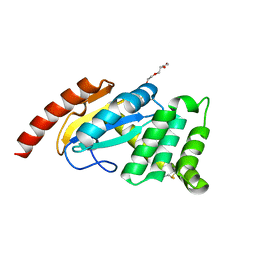 | |
3RXS
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine (F04 and A06, cocktail experiment) | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
4MI8
 
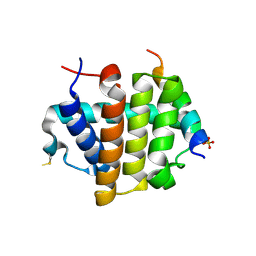 | |
3F51
 
 | | Crystal Structure of the clp gene regulator ClgR from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
4LOA
 
 | | X-ray structure of the de-novo design amidase at the resolution 1.8A, Northeast Structural Genomics Consortium (NESG) Target OR398 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, De-novo design amidase | | Authors: | Kuzin, A, Lew, S, Vorobiev, S.M, Seetharaman, J, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Khersonsky, O, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR398
To be Published
|
|
4LSA
 
 | | Crystal structure of BRI1 sud1 (Gly643Glu) bound to brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, Protein BRASSINOSTEROID INSENSITIVE 1, ... | | Authors: | Santiago, J, Henzler, C, Hothorn, M. | | Deposit date: | 2013-07-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases.
Science, 341, 2013
|
|
1IS9
 
 | | Endoglucanase A from Clostridium thermocellum at atomic resolution | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, endoglucanase A | | Authors: | Schmidt, A, Gonzalez, A, Morris, R.J, Costabel, M, Alzari, P.M, Lamzin, V.S. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Advantages of high-resolution phasing: MAD to atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3F4T
 
 | |
4KX6
 
 | | Plasticity of the quinone-binding site of the complex II homolog quinol:fumarate reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Singh, P.K, Sarwar, M, Maklashina, E, Kotlyar, V, Rajagukguk, S, Tomasiak, T.M, Cecchini, G, Iverson, T.M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasticity of the Quinone-binding Site of the Complex II Homolog Quinol:Fumarate Reductase.
J.Biol.Chem., 288, 2013
|
|
3RYH
 
 | | GMPCPP-Tubulin: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Nawrotek, A, Knossow, M, Gigant, B. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Determinants That Govern Microtubule Assembly from the Atomic Structure of GTP-Tubulin.
J.Mol.Biol., 412, 2011
|
|
3F5N
 
 | |
1IT2
 
 | | Hagfish deoxy hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin | | Authors: | Mito, M, Chong, K.T, Park, S.-Y, Tame, J.R. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of deoxy- and carbonmonoxyhemoglobin F1 from the hagfish Eptatretus burgeri
J.Biol.Chem., 277, 2002
|
|
1ITO
 
 | | Crystal Structure Analysis of Bovine Spleen Cathepsin B-E64c complex | | Descriptor: | Cathepsin B, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE | | Authors: | Yamamoto, A, Tomoo, T, Matsugi, K, Hara, T, In, Y, Murata, M, Kitamura, K, Ishida, T. | | Deposit date: | 2002-01-19 | | Release date: | 2003-01-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Structural basis for development of cathepsin B-specific noncovalent-type inhibitor: crystal structure of cathepsin B-E64c complex
BIOCHIM.BIOPHYS.ACTA, 1597, 2002
|
|
3S9H
 
 | | RB69 DNA Polymerase Triple Mutant(L561A/S565G/Y567A) ternary complex with dUpNpp and a dideoxy-terminated primer in the presence of Ca2+ | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-06-01 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase.
Biochemistry, 50, 2011
|
|
3S1A
 
 | | Crystal structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.W, Rossi, G, Weigand, S, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2011-05-14 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Combined SAXS/EM Based Models of the S. elongatus Post-Translational Circadian Oscillator and its Interactions with the Output His-Kinase SasA.
Plos One, 6, 2011
|
|
3F7K
 
 | | X-ray Crystal Structure of an Alvinella pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper,Zinc Superoxide Dismutase, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
4L31
 
 | | Tankyrase 2 in complex with methyl 4-(4-oxochromen-2-yl)benzoate | | Descriptor: | GLYCEROL, SULFATE ION, Tankyrase-2, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2013-06-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of tankyrase inhibiting flavones with increased potency and isoenzyme selectivity.
J.Med.Chem., 56, 2013
|
|
1IUO
 
 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant complexed with acetates | | Descriptor: | ACETATE ION, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
4LRZ
 
 | | Crystal Structure of the E.coli DhaR(N)-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-dependent dihydroxyacetone kinase operon regulatory protein, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
5REE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2217052426 | | Descriptor: | (2R,3R)-1-benzyl-2-methylpiperidin-3-ol, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102395 | | Descriptor: | 2-[(4-acetylpiperazin-1-yl)sulfonyl]benzonitrile, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3SCK
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus civet strain complexed with human-civet chimeric receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2 chimera, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
1J9A
 
 | | OLIGORIBONUCLEASE | | Descriptor: | OLIGORIBONUCLEASE, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Ladner, J.E, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-24 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of Haemophilus Influenzae HI1715, an Oligoribonuclease
To be Published
|
|
