4I2H
 
 | | Ternary complex of mouse TdT with ssDNA and AMPcPP | | Descriptor: | 5'-D(P*AP*AP*AP*AP*A)-3', DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA nucleotidylexotransferase, ... | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Intermediates along the Catalytic Cycle of Terminal Deoxynucleotidyltransferase: Dynamical Aspects of the Two-Metal Ion Mechanism.
J.Mol.Biol., 425, 2013
|
|
4I86
 
 | | Crystal structure of PilZ domain of CeSA from cellulose synthesizing bacterium | | Descriptor: | Cellulose synthase 1 | | Authors: | Fujiwara, T, Komoda, K, Sakurai, N, Tanaka, I, Yao, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A
Biochem.Biophys.Res.Commun., 431, 2013
|
|
2AEG
 
 | | X-Ray Crystal Structure of Protein Atu5096 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR63. | | Descriptor: | hypothetical protein AGR_pAT_140 | | Authors: | Forouhar, F, Abashidze, M, Kuzin, A.P, Vorobiev, S.M, Shastry, R, Cooper, B, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-22 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Hypothetical Protein Atu5096 from Agrobacterium tumefaciens.
To be Published
|
|
3OIP
 
 | | Crystal structure of Yeast telomere protein Cdc13 OB1 | | Descriptor: | Cell division control protein 13 | | Authors: | Sun, J, Yang, Y, Wan, K, Mao, N, Yu, T.Y, Lin, Y.C, DeZwaan, D.C, Freeman, B.C, Lin, J.J, Lue, N.F, Lei, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha.
Cell Res., 21, 2011
|
|
4I53
 
 | |
2AG0
 
 | |
2WP1
 
 | | Structure of Brdt bromodomain 2 bound to an acetylated histone H3 peptide | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H3 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
2WW4
 
 | | a triclinic crystal form of E. coli 4-diphosphocytidyl-2C-methyl-D- erythritol kinase | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL | | Authors: | Kalinowska-Tluscik, J, Miallau, L, Gabrielsen, M, Leonard, G.A, McSweeney, S.M, Hunter, W.N. | | Deposit date: | 2009-10-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Triclinic Crystal Form of Escherichia Coli 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase and Reassessment of the Quaternary Structure.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
3OJ2
 
 | |
4I2E
 
 | | Ternary complex of mouse TdT with ssDNA and AMPcPP | | Descriptor: | 5'-D(*AP*AP*AP*AP*A)-3', COBALT (II) ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Intermediates along the Catalytic Cycle of Terminal Deoxynucleotidyltransferase: Dynamical Aspects of the Two-Metal Ion Mechanism.
J.Mol.Biol., 425, 2013
|
|
5BYI
 
 | |
2A14
 
 | | Crystal Structure of Human Indolethylamine N-methyltransferase with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, indolethylamine N-methyltransferase | | Authors: | Dong, A, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Human Indolethylamine
N-methyltransferase in complex with SAH.
To be Published
|
|
3I33
 
 | | Crystal structure of the human 70kDa heat shock protein 2 (Hsp70-2) ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Heat shock-related 70 kDa protein 2, ... | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Kotzsch, A, Nielsen, T.K, Nordlund, P, Nyman, T, Moche, M, Persson, C, Roos, A.K, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Weigelt, J, Welin, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
2X1E
 
 | | The crystal structure of mature acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum in complex 6- aminopenicillanic acid | | Descriptor: | (2S,5R,6R)-6-AMINO-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, ACYL-COENZYME, CHLORIDE ION, ... | | Authors: | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
2X4S
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a peptide representing the epitope of the H5N1 (Avian Flu) Nucleoprotein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals
J.Am.Chem.Soc., 131, 2009
|
|
2A43
 
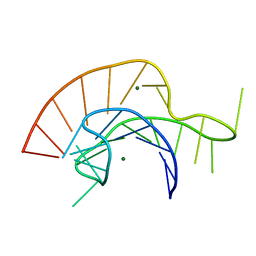 | | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif | | Descriptor: | MAGNESIUM ION, RNA Pseudoknot | | Authors: | Pallan, P.S, Marshall, W.S, Harp, J, Jewett III, F.C, Wawrzak, Z, Brown II, B.A, Rich, A, Egli, M. | | Deposit date: | 2005-06-27 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Biochemistry, 44, 2005
|
|
4I2J
 
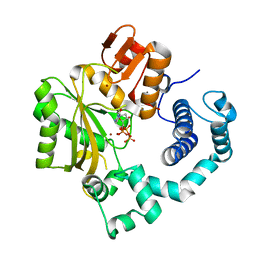 | | Binary complex of mouse TdT with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA nucleotidylexotransferase, ZINC ION | | Authors: | Gouge, J, Delarue, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Intermediates along the Catalytic Cycle of Terminal Deoxynucleotidyltransferase: Dynamical Aspects of the Two-Metal Ion Mechanism.
J.Mol.Biol., 425, 2013
|
|
5C17
 
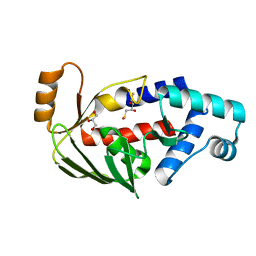 | | Crystal structure of the mercury-bound form of MerB2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-13 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
2X7O
 
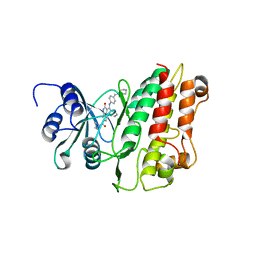 | | Crystal structure of TGFbRI complexed with an indolinone inhibitor | | Descriptor: | (3Z)-N-ETHYL-N-METHYL-2-OXO-3-(PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]AMINO}METHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-6-CARBOXAMIDE, TGF-BETA RECEPTOR TYPE I | | Authors: | Roth, G.J, Heckel, A, Brandl, T, Grauert, M, Hoerer, S, Kley, J.T, Schnapp, G, Baum, P, Mennerich, D, Schnapp, A, Park, J.E. | | Deposit date: | 2010-03-03 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis and Evaluation of Indolinones as Inhibitors of the Transforming Growth Factor Beta Receptor I (Tgfbri)
J.Med.Chem., 53, 2010
|
|
4I6J
 
 | | A ubiquitin ligase-substrate complex | | Descriptor: | Cryptochrome-2, F-box/LRR-repeat protein 3, S-phase kinase-associated protein 1 | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
3P35
 
 | |
5C1I
 
 | | m1A58 tRNA methyltransferase mutant - D170A | | Descriptor: | SULFATE ION, tRNA (adenine(58)-N(1))-methyltransferase TrmI | | Authors: | Ponchon, L, Degut, C, Folly-Klan, M, Barraud, P, Tisne, C. | | Deposit date: | 2015-06-14 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The m1A58 modification in eubacterial tRNA: An overview of tRNA recognition and mechanism of catalysis by TrmI.
Biophys.Chem., 210, 2016
|
|
3P4Q
 
 | | Crystal structure of Menaquinol:oxidoreductase in complex with oxaloacetate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
4I76
 
 | | Crystal structure of transcriptional regulator TM1030 with octanol | | Descriptor: | 1,2-ETHANEDIOL, OCTAN-1-OL, Transcriptional regulator, ... | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of transcriptional regulator TM1030 with octanol
To be Published
|
|
