3MJF
 
 | | Phosphoribosylamine-glycine ligase from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystal structure of phosphoribosylamine-glycine ligase from Yersinia pestis.
To be Published
|
|
8BK8
 
 | |
8BLC
 
 | | Structure of the GroEL-ATP complex plunge-frozen 50 ms after mixing with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, MAGNESIUM ION, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
2V3R
 
 | | Hypocrea jecorina Cel7A in complex with (S)-dihydroxy-phenanthrenolol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(2S)-2-HYDROXY-3-(9-PHENANTHRYLOXY)PROPYL]AMINO}PROPANE-1,3-DIOL, COBALT (II) ION, ... | | Authors: | Fagerstrom, A, Sandgren, M, Berg, U, Stahlberg, J. | | Deposit date: | 2007-06-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Study of the Chiral Recognition Mechanisms of Cellobiohydrolase Cel7A for Ligands Based on the Beta-Blocker Motif: Crystal Structures, Microcalorimetry and Computational Modelling of Cel7A-Inhibitor Complexes.
To be Published
|
|
2V4C
 
 | |
3MNM
 
 | | Crystal structure of GAE domain of GGA2p from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-binding protein GGA2, GLYCEROL, SULFATE ION | | Authors: | Fang, P, Wang, J, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-08 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for the specificity of the GAE domain of yGGA2 for its accessory proteins Ent3 and Ent5
Biochemistry, 49, 2010
|
|
8B7B
 
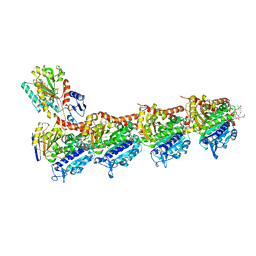 | | Tubulin - maytansinoid - 6 complex | | Descriptor: | (~{Z})-11-[[(1~{R},2~{R},3~{S},5~{S},6~{S},16~{E},18~{E},20~{R},21~{S})-11-chloranyl-12,20-dimethoxy-2,5,9,16-tetramethyl-21-oxidanyl-8,23-bis(oxidanylidene)-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1^{10,14}.0^{3,5}]hexacosa-10(26),11,13,16,18-pentaen-6-yl]oxy]-11-oxidanylidene-undec-2-enoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, J.F, Steinmetz, M.O, Prota, A.E, Pieraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
4WRX
 
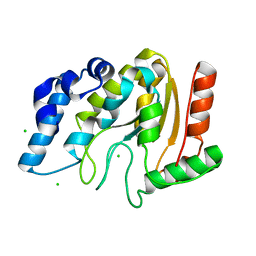 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form V | | Descriptor: | CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
4WS0
 
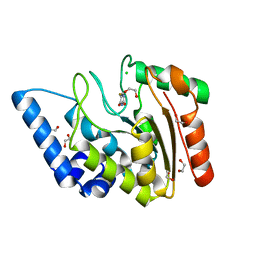 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (A), Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS3
 
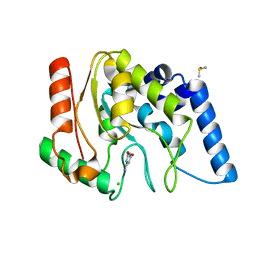 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 6-aminouracil, Form IV | | Descriptor: | 6-aminopyrimidine-2,4(3H,5H)-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8B7A
 
 | | Tubulin - maytansinoid - 4 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Boiarska, Z, Perez-Pena, H, Abel, A.-C, Marzullo, P, Alvarez-Bernad, B, Bonato, F, Santini, B, Horvath, D, Lucena-Agell, D, Vasile, F, Sironi, M, Diaz, F.J, Steinmetz, M.O, Prota, A.E, Piaraccini, S, Passarella, D. | | Deposit date: | 2022-09-29 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Chemistry, 29, 2023
|
|
1XR9
 
 | | Crystal Structures of HLA-B*1501 in Complex with Peptides from Human UbcH6 and Epstein-Barr Virus EBNA-3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G, Blicher, T, Johannessen, B.R, Kristensen, O, Buus, S, Gajhede, M. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3MP8
 
 | | Crystal structure of Sgf29 tudor domain | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, ACETIC ACID, GLYCEROL, ... | | Authors: | Li, J, Wu, M, Ruan, J, Zang, J. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
1XRQ
 
 | | Crystal structure of active site F1-mutant E245Q soaked with peptide Phe-Leu | | Descriptor: | LEUCINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
8B5K
 
 | | Structure of haloalkane dehalogenase DmmarA from Mycobacterium marinum at pH 6.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Snajdarova, K, Marek, M. | | Deposit date: | 2022-09-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Atypical homodimerization revealed by the structure of the (S)-enantioselective haloalkane dehalogenase DmmarA from Mycobacterium marinum.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1XMY
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With (R)-Rolipram | | Descriptor: | MAGNESIUM ION, ROLIPRAM, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
3LZN
 
 | | Crystal Structure Analysis of the apo P19 protein from Campylobacter jejuni at 1.59 A at pH 9 | | Descriptor: | P19 protein, SULFATE ION, ZINC ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
3LZV
 
 | | Structure of Nelfinavir-resistant HIV-1 protease (D30N/N88D) in complex with Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 Protease, ... | | Authors: | Schiffer, C.A, Kolli, M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
4WS8
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 2-thiouracil, Form V | | Descriptor: | 2-thioxo-2,3-dihydropyrimidin-4(1H)-one, CHLORIDE ION, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7ZNY
 
 | | Cryo-EM structure of the canine distemper virus tetrameric attachment glycoprotein | | Descriptor: | Hemagglutinin glycoprotein | | Authors: | Kalbermatter, D, Jeckelmann, J.-M, Wyss, M, Plattet, P, Fotiadis, D. | | Deposit date: | 2022-04-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and supramolecular organization of the canine distemper virus attachment glycoprotein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3ELB
 
 | | Human CTP: Phosphoethanolamine Cytidylyltransferase in complex with CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Ethanolamine-phosphate cytidylyltransferase, GLYCEROL | | Authors: | Karlberg, T, Welin, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wikstrom, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-22 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human CTP:Phosphoethanolamine Cytidylyltransferase
To be Published
|
|
3MRK
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with AFP137 nonapeptide | | Descriptor: | 9-meric peptide from Alpha-fetoprotein, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of MHC class I HLA-A2 molecule complexed with AFP137 nonapeptide
To be Published
|
|
2VRE
 
 | | crystal structure of human peroxisomal delta3,5,delta2,4-dienoyl coa isomerase | | Descriptor: | CHLORIDE ION, DELTA(3,5)-DELTA(2,4)-DIENOYL-COA ISOMERASE | | Authors: | Yue, W, Guo, K, von Delft, F, Pilka, E, Murray, J, Roos, A, Kochan, G, Bountra, C, Arrowsmith, C, Wikstrom, M, Edwards, A, Oppermann, U. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Peroxisomal Delta3,5, Delta2,4-Dienoyl Coa Isomerase (Ech1)
To be Published
|
|
3ZJY
 
 | | Crystal Structure of Importin 13 - RanGTP - eIF1A complex | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 1A, X-CHROMOSOMAL, GTP-BINDING NUCLEAR PROTEIN RAN, ... | | Authors: | Gruenwald, M, Lazzaretti, D, Bono, F. | | Deposit date: | 2013-01-21 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis for the Nuclear Export Activity of Importin13.
Embo J., 32, 2013
|
|
