409D
 
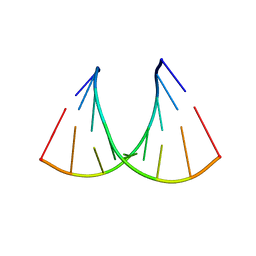 | | CRYSTAL STRUCTURE OF AN RNA R(CCCIUGGG) WITH THREE INDEPENDENT DUPLEXES INCORPORATING TANDEM I.U WOBBLES | | Descriptor: | RNA (5'-R(*CP*CP*CP*IP*UP*GP*GP*G)-3') | | Authors: | Pan, B, Mitra, S.N, Sun, L, Hart, D, Sundaralingam, M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an RNA octamer duplex r(CCCIUGGG)2 incorporating tandem I.U wobbles.
Nucleic Acids Res., 26, 1998
|
|
418D
 
 | |
4A2M
 
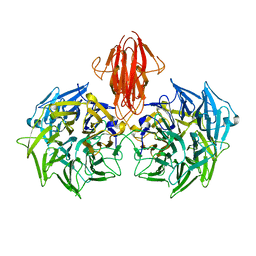 | | Structure of the periplasmic domain of the heparin and heparan sulphate sensing hybrid two component system BT4663 in apo and ligand bound forms | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, TWO-COMPONENT SYSTEM SENSOR HISTIDINE KINASE/RESPONSE | | Authors: | Lowe, E.C, Basle, A, Czjzek, M, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A Scissor Blade-Like Closing Mechanism Implicated in Transmembrane Signaling in a Bacteroides Hybrid Two-Component System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
410D
 
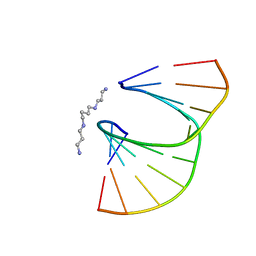 | | DUPLEX [5'-D(GCGTA+TACGC)]2 WITH INCORPORATED 2'-O-ETHOXYMETHYLENE RIBONUCLEOSIDE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(T38)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Tereshko, V, Portmann, S, Tay, E.C, Martin, P, Natt, F, Altmann, K.H, Egli, M. | | Deposit date: | 1998-06-30 | | Release date: | 1998-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Correlating structure and stability of DNA duplexes with incorporated 2'-O-modified RNA analogues.
Biochemistry, 37, 1998
|
|
437D
 
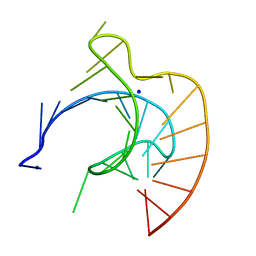 | | CRYSTAL STRUCTURE OF AN RNA PSEUDOKNOT FROM BEET WESTERN YELLOW VIRUS INVOLVED IN RIBOSOMAL FRAMESHIFTING | | Descriptor: | MAGNESIUM ION, RNA PSEUDOKNOT, SODIUM ION | | Authors: | Su, L, Chen, L, Egli, M, Berger, J.M, Rich, A. | | Deposit date: | 1998-11-24 | | Release date: | 1998-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot.
Nat.Struct.Biol., 6, 1999
|
|
4A93
 
 | | RNA Polymerase II elongation complex containing a CPD Lesion | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP*CP*T*TTP*TP*TP*CP*C BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP*AP)-3', ... | | Authors: | Walmacq, C, Cheung, A.C.M, Kireeva, M.L, Lubkowska, L, Ye, C, Gotte, D, Strathern, J.N, Carell, T, Cramer, P, Kashlev, M. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Translesion Transcription by RNA Polymerase II and its Role in Cellular Resistance to DNA Damage.
Mol.Cell, 46, 2012
|
|
4A6L
 
 | | beta-tryptase inhibitor | | Descriptor: | 1-{3-[1-({5-[(2-fluorophenyl)ethynyl]furan-2-yl}carbonyl)piperidin-4-yl]phenyl}methanamine, TRYPTASE ALPHA/BETA-1 | | Authors: | Mathieu, M, Maignan, S. | | Deposit date: | 2011-11-04 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Library Design and the Discovery of a Potent and Selective Mast Cell Beta-Tryptase Inhibitor as an Oral Therapeutic Agent.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7RPV
 
 | |
4AC7
 
 | | The crystal structure of Sporosarcina pasteurii urease in complex with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, HYDROXIDE ION, ... | | Authors: | Benini, S, Kosikowska, P, Cianci, M, Gonzalez Vara, A, Berlicki, L, Ciurli, S. | | Deposit date: | 2011-12-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of Sporosarcina Pasteurii Urease in a Complex with Citrate Provides New Hints for Inhibitor Design.
J.Biol.Inorg.Chem., 18, 2013
|
|
6UD7
 
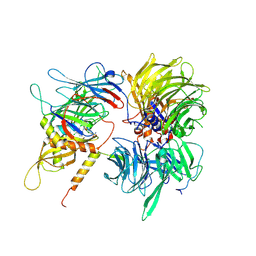 | | Crystal structure of full-length human DCAF15-DDB1(deltaBPB)-DDA1-RBM39 in complex with indisulam | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1,DNA damage-binding protein 1, ... | | Authors: | Bussiere, D.E, Shu, W, Xie, L, Knapp, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
4A0O
 
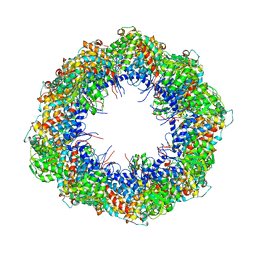 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4WR5
 
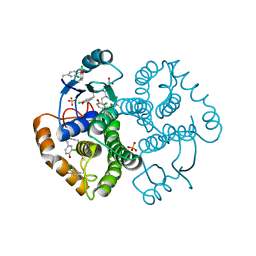 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4A27
 
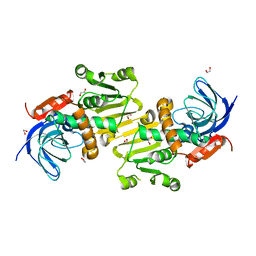 | | Crystal structure of human synaptic vesicle membrane protein VAT-1 homolog-like protein | | Descriptor: | 1,2-ETHANEDIOL, SYNAPTIC VESICLE MEMBRANE PROTEIN VAT-1 HOMOLOG-LIKE | | Authors: | Vollmar, M, Shafqat, N, Muniz, J.R.C, Krojer, T, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Synaptic Vesicle Membrane Protein Vat-1 Homolog-Like Protein
To be Published
|
|
4AR7
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2012-04-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structure of a Fluorescent Protein from Aequorea Victoria Bearing the Obligate-Monomer Mutation A206K.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4AAQ
 
 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4ARG
 
 | | Structure of the immature retroviral capsid at 8A resolution by cryo- electron microscopy | | Descriptor: | M-PMV DPRO CANC PROTEIN | | Authors: | Bharat, T.A.M, Davey, N.E, Ulbrich, P, Riches, J.D, Marco, A.D, Rumlova, M, Sachse, C, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structure of the Immature Retroviral Capsid at 8A Resolution by Cryo-Electron Microscopy.
Nature, 487, 2012
|
|
4A9Z
 
 | | CRYSTAL STRUCTURE OF HUMAN P63 TETRAMERIZATION DOMAIN | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, TUMOR PROTEIN 63 | | Authors: | Muniz, J.R.C, Coutandin, D, Salah, E, Chaikuad, A, Vollmar, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, von Delft, F, Knapp, S. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of Human P63 Tetramerization Domain
To be Published
|
|
4AHW
 
 | |
8UAK
 
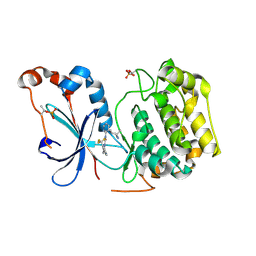 | | Crystal structure of the catalytic domain of human PKC alpha (D463N, V568I, S657E) in complex with Darovasertib (NVP-LXS196) at 2.82-A resolution | | Descriptor: | (6M)-3-amino-N-[3-(4-amino-4-methylpiperidin-1-yl)pyridin-2-yl]-6-[3-(trifluoromethyl)pyridin-2-yl]pyrazine-2-carboxamide, Protein kinase C alpha type | | Authors: | Romanowski, M.J, Lam, J, Visser, M. | | Deposit date: | 2023-09-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of Darovasertib (NVP-LXS196), a Pan-PKC Inhibitor for the Treatment of Metastatic Uveal Melanoma.
J.Med.Chem., 67, 2024
|
|
7RLU
 
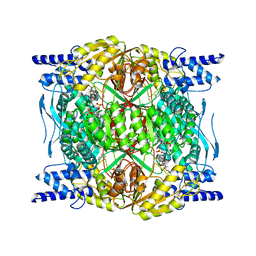 | | Structure of ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) in complex with NADP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
7RLT
 
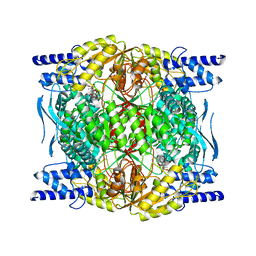 | | Structure of ligand-free ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase | | Authors: | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
6U5F
 
 | | CryoEM Structure of Pyocin R2 - precontracted - collar | | Descriptor: | Collar PA0615, Sheath PA0622, Tube PA0623 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
6UA1
 
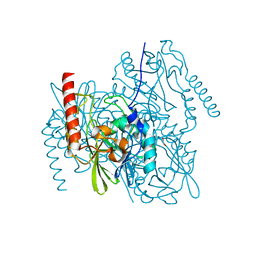 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form | | Descriptor: | 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase) | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form.
To Be Published
|
|
6UAF
 
 | | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem
To Be Published
|
|
4TO7
 
 | |
