5BSG
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NADP+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5BTS
 
 | |
7D54
 
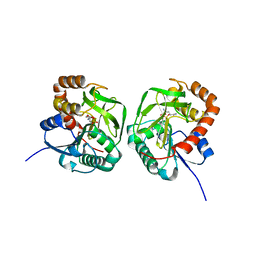 | | Crstal structure MsGATase with Gln | | Descriptor: | GLUTAMINE, Glutamine amidotransferase class-I | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5AQA
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | OFF7_DB04V3, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5AUR
 
 | | Hydrogenobacter thermophilus cytochrome c552 dimer formed by domain swapping at N-terminal region | | Descriptor: | Cytochrome c-552, HEME C, IODIDE ION | | Authors: | Ren, C, Nagao, S, Yamanaka, M, Kamikubo, H, Komori, H, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Oligomerization enhancement and two domain swapping mode detection for thermostable cytochrome c552via the elongation of the major hinge loop.
Mol Biosyst, 11, 2015
|
|
5AV2
 
 | |
1SGU
 
 | | Comparing the Accumulation of Active Site and Non-active Site Mutations in the HIV-1 Protease | | Descriptor: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, POL polyprotein | | Authors: | Clemente, J.C, Moose, R.E, Hemrajani, R, Govindasamy, L, Reutzel, R, Mckenna, R, Abandje-McKenna, M, Goodenow, M.M, Dunn, B.M. | | Deposit date: | 2004-02-24 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparing the Accumulation of Active- and Nonactive-Site Mutations in the HIV-1 Protease.
Biochemistry, 43, 2004
|
|
7D4R
 
 | | SpuA native structure | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DMO
 
 | |
7D53
 
 | | SpuA mutant - H221N with Glu | | Descriptor: | GLUTAMIC ACID, MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DBT
 
 | | Crystal structure of catalytic domain of Anhydrobiosis-related Mn-dependent Peroxidase (AMNP) from Ramazzottius varieornatus (Mn2+-bound form) | | Descriptor: | AMNP/g12777, MANGANESE (II) ION | | Authors: | Yoshida, Y, Satoh, T, Ota, C, Tanaka, S, Horikawa, D.D, Tomita, M, Kato, K, Arakawa, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-series transcriptomic screening of factors contributing to the cross-tolerance to UV radiation and anhydrobiosis in tardigrades.
Bmc Genomics, 23, 2022
|
|
7DBU
 
 | | Crystal structure of catalytic domain of Anhydrobiosis-related Mn-dependent Peroxidase (AMNP) from Ramazzottius varieornatus (Zn2+-bound form) | | Descriptor: | AMNP/g12777, ZINC ION | | Authors: | Yoshida, Y, Satoh, T, Ota, C, Tanaka, S, Horikawa, D.D, Tomita, M, Kato, K, Arakawa, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-series transcriptomic screening of factors contributing to the cross-tolerance to UV radiation and anhydrobiosis in tardigrades.
Bmc Genomics, 23, 2022
|
|
5B1X
 
 | |
7D50
 
 | | SpuA mutant - H221N with glutamyl-thioester | | Descriptor: | MAGNESIUM ION, Probable glutamine amidotransferase | | Authors: | Chen, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure and mechanism of the gamma-glutamyl-gamma-aminobutyrate hydrolase SpuA from Pseudomonas aeruginosa.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3VSD
 
 | | Crystal Structure of the K127A Mutant of O-Phosphoserine Sulfhydrylase Complexed with External Schiff Base of Pyridoxal 5'-Phosphate with O-Acetyl-L-Serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, O-ACETYLSERINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
7DT3
 
 | | Crystal structure of human transthyretin in complex with 4-chloro-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid | | Descriptor: | 4-chloranyl-9,10-bis(oxidanylidene)anthracene-2-carboxylic acid, CALCIUM ION, Transthyretin | | Authors: | Kitakami, R, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Inhibitory activities of anthraquinone and xanthone derivatives against transthyretin amyloidogenesis.
Bioorg.Med.Chem., 44, 2021
|
|
5AQ7
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | D12_DB04V3, MALONATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
7DT5
 
 | |
7DT6
 
 | |
7DT8
 
 | |
7D5Z
 
 | | Crystal structure of EBV gH/gL bound with neutralizing antibody 1D8 | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, heavy chain of 1D8, ... | | Authors: | Zhu, Q, Shan, S, Yu, J, Wang, X, Zhang, L, Zeng, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | A Neutralizing Antibody Targeting a New Site of Vulnerability on Epstein-Barr Virus gH/gL Protects against Dual-Tropic Infection
To Be Published
|
|
3O8V
 
 | | Crystal Structure of the Tudor Domains from FXR1 | | Descriptor: | Fragile X mental retardation syndrome-related protein 1 | | Authors: | Lam, R, Guo, Y.H, Adams-Cioaba, M, Bian, C.B, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2.
Plos One, 5, 2010
|
|
7DMN
 
 | | Crystal structure of two pericyclases catalyzing [4+2] cycloaddition | | Descriptor: | Diels-Alderase fsa2, GLYCEROL | | Authors: | Chi, C.B, Wang, Z.D, Liu, T, Zhang, Z.Y, Ma, M. | | Deposit date: | 2020-12-04 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Fsa2 and Phm7 Catalyzing [4 + 2] Cycloaddition Reactions with Reverse Stereoselectivities in Equisetin and Phomasetin Biosynthesis.
Acs Omega, 6, 2021
|
|
5DKI
 
 | | Yeast 20S proteasome in complex with alkyne-PI | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Beck, P, Cui, H, Groll, M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted Delivery of Proteasome Inhibitors to Somatostatin-Receptor-Expressing Cancer Cells by Octreotide Conjugation.
Chemmedchem, 10, 2015
|
|
