5TSG
 
 | | PilB from Geobacter metallireducens bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | McCallum, M, Tammam, S, Khan, A, Burrows, L, Howell, P.L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4011 Å) | | Cite: | The molecular mechanism of the type IVa pilus motors.
Nat Commun, 8, 2017
|
|
5FIN
 
 | | DARPins as a new tool for experimental phasing in protein crystallography | | Descriptor: | MERCURY (II) ION, NI3C DARPIN MUTANT5 HG-SITE N1 | | Authors: | Batyuk, A, Honegger, A, Andres, F, Briand, C, Gruetter, M, Plueckthun, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Darpins as a New Tool for Experimental Phasing in Protein Crystallography
To be Published
|
|
5FSN
 
 | | MTH1 substrate recognition: Complex with a aminomethylpyrimidinyl oxypropanol. | | Descriptor: | 3-(2-AMINO-6-METHYL-PYRIMIDIN-4-YL)OXYPROPAN-1-OL, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
7Q7Z
 
 | |
6F4Y
 
 | |
2M84
 
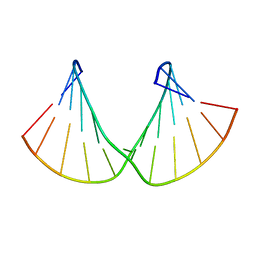 | | Structure of 2'F-RNA/2'F-ANA chimeric duplex | | Descriptor: | 2'F-RNA/2'F-ANA CHIMERIC DUPLEX | | Authors: | Martin-Pintado, N, Deleavey, G, Portella, G, Campos, R, Orozco, M, Damha, M, Gonzalez, C. | | Deposit date: | 2013-05-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone FCHO Hydrogen Bonds in 2'F-Substituted Nucleic Acids.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7Q81
 
 | |
6F54
 
 | |
7QDW
 
 | |
5O6Y
 
 | | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide, ... | | Authors: | Fadouloglou, V.E, Kotsifaki, D, Kokkinidis, M. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of the Bc1960 peptidoglycan N-acetylglucosamine deacetylase in complex with 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide
To Be Published
|
|
8C8B
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 48). | | Descriptor: | 4-[[(2~{S})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-~{N}-prop-2-enyl-quinazoline-2-carboxamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21).
To Be Published
|
|
1BK4
 
 | | CRYSTAL STRUCTURE OF RABBIT LIVER FRUCTOSE-1,6-BISPHOSPHATASE AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEIN (FRUCTOSE-1,6-BISPHOSPHATASE), SULFATE ION | | Authors: | Ghosh, D, Weeks, C.M, Erman, M, Roszak, A.W, Kaiser, R, Jornvall, H. | | Deposit date: | 1998-07-14 | | Release date: | 1998-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rabbit liver fructose 1,6-bisphosphatase at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8CD3
 
 | |
5IYT
 
 | | Complex structure of EV-B93 main protease 3C with N-Ethyl 4-((1-cycloheptyl-1,2-dihydropyrazol-3-one-5-yl)-amino)-4-oxo-2Z-butenamide | | Descriptor: | EV-B93 main protease 3C, N-Ethyl 4-((1-cycloheptyl-1,2-dihydropyrazol-3-one-5-yl)-amino)-4-oxo-butanamide | | Authors: | Kaczmarska, Z, Becker, D, Rademann, J, Coll, M. | | Deposit date: | 2016-03-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding fragments.
Nat Commun, 7, 2016
|
|
2ZX1
 
 | | Rhamnose-binding lectin CSL3 | | Descriptor: | CSL3, PHOSPHATE ION | | Authors: | Shirai, T, Watababe, Y, Lee, M, Ogawa, T, Muramoto, K. | | Deposit date: | 2008-12-19 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhamnose-binding lectin CSL3: unique pseudo-tetrameric architecture of a pattern recognition protein
J.Mol.Biol., 391, 2009
|
|
5TV4
 
 | | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, L-glycero-alpha-D-manno-heptopyranose-(1-7)-L-glycero-alpha-D-manno-heptopyranose-(1-3)-L-glycero-alpha-D-manno-heptopyranose-(1-5)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-alpha-D-glucopyranose-(1-6)-2-amino-2-deoxy-alpha-D-glucopyranose, LAURIC ACID, ... | | Authors: | Mi, W, Walz, T, Liao, M. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of MsbA-mediated lipopolysaccharide transport.
Nature, 549, 2017
|
|
5FVJ
 
 | | Crystal structure of TacT (tRNA acetylating toxin) from Salmonella | | Descriptor: | ACETYL COENZYME *A, PUTATIVE ACETYLTRANSFERASE | | Authors: | Przydacz, M, Wong, C.T, Cheverton, A.M, Gollan, B, Mylona, A, Helaine, S, Hare, S.A. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Salmonella Toxin Promotes Persister Formation Through Acetylation of tRNA.
Mol.Cell, 63, 2016
|
|
1BNR
 
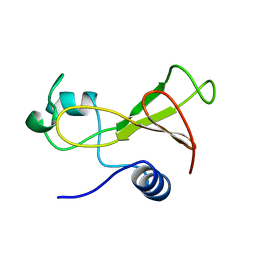 | | BARNASE | | Descriptor: | BARNASE (G SPECIFIC ENDONUCLEASE) | | Authors: | Bycroft, M. | | Deposit date: | 1995-03-31 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional solution structure of barnase using nuclear magnetic resonance spectroscopy.
Biochemistry, 30, 1991
|
|
5FRP
 
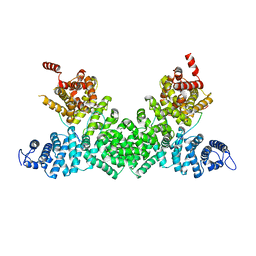 | | Structure of the Pds5-Scc1 complex and implications for cohesin function | | Descriptor: | MCD1-LIKE PROTEIN, SISTER CHROMATID COHESION PROTEIN PDS5 | | Authors: | Muir, K.W, Kschonsak, M, Li, Y, Metz, J, Haering, C.H, Panne, D. | | Deposit date: | 2015-12-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure of the Pds5-Scc1 Complex and Implications for Cohesin Function
Cell Rep., 14, 2016
|
|
7QTO
 
 | |
1BZG
 
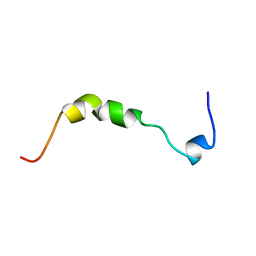 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE-RELATED PROTEIN (1-34) IN NEAR-PHYSIOLOGICAL SOLUTION, NMR, 30 STRUCTURES | | Descriptor: | PARATHYROID HORMONE-RELATED PROTEIN | | Authors: | Weidler, M, Marx, U.C, Seidel, G, Roesch, P. | | Deposit date: | 1998-10-28 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of human parathyroid hormone-related protein(1-34) in near-physiological solution.
FEBS Lett., 444, 1999
|
|
5GSS
 
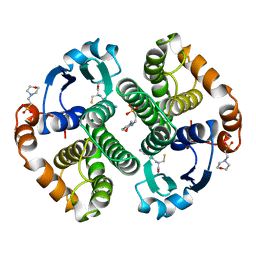 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-11 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
7QTP
 
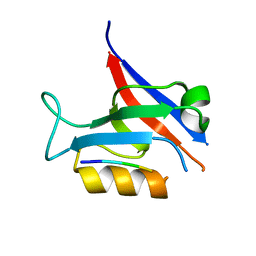 | |
7QBP
 
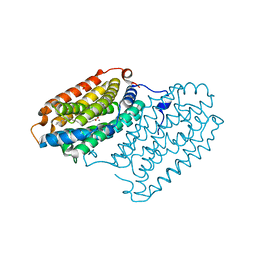 | | Crystal structure of R2-like ligand-binding oxidase from Saccharopolyspora Erythraea | | Descriptor: | FE (III) ION, MANGANESE (III) ION, PALMITIC ACID, ... | | Authors: | Srinivas, V, Diamanti, R, Lebrette, H, Hogbom, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Comparative structural analysis provides new insights into the function of R2-like ligand-binding oxidase.
Febs Lett., 596, 2022
|
|
5G3L
 
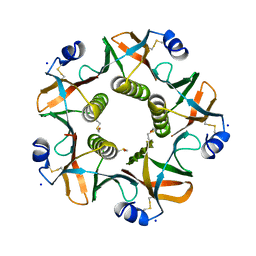 | | ESCHERICHIA COLI HEAT LABILE ENTEROTOXIN TYPE IIB B-PENTAMER COMPLEXED WITH SIALYLATED SUGAR | | Descriptor: | HEAT-LABILE ENTEROTOXIN IIB, B CHAIN, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Zalem, D, Benktander, J, Ribeiro, J.P, Varrot, A, Lebens, M, Imberty, A, Teneberg, S. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Biochemical and Structural Characterization of the Novel Sialic Acid-Binding Site of Escherichia Coli Heat-Labile Enterotoxin Lt-Iib.
Biochem.J., 473, 2016
|
|
