2PCF
 
 | | THE COMPLEX OF CYTOCHROME F AND PLASTOCYANIN DETERMINED WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | Descriptor: | COPPER (II) ION, CYTOCHROME F, HEME C, ... | | Authors: | Ubbink, M, Ejdeback, M, Karlsson, B.G, Bendall, D.S. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics.
Structure, 6, 1998
|
|
6WRL
 
 | | The interaction of chlorido(1,5-cyclooctadiene)([4-(2-((tert-butoxycarbonyl)amino)-3-methoxy-3-oxopropyl)-1,3-dimethyl-1H-imidazol-3-ide])rhodium(I) with HEWL after 1 week | | Descriptor: | ACETATE ION, Lysozyme, Rhodium, ... | | Authors: | Sullivan, M.P, John, M, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-04-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Combined Spectroscopic and Protein Crystallography Study Reveals Protein Interactions of Rh I (NHC) Complexes at the Molecular Level.
Inorg.Chem., 59, 2020
|
|
4ZWN
 
 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
5K98
 
 | | Structure of HipA-HipB-O2-O3 complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*CP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*G)-3'), ... | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
6OOO
 
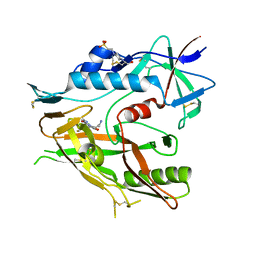 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-IV-226. | | Descriptor: | (3S,5R)-5-amino-N~3~-(4-chloro-3-fluorophenyl)-N~1~-propylpiperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-23 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of Small Molecules That Sensitize HIV-1 Infected Cells to Antibody-Dependent Cellular Cytotoxicity.
Acs Med.Chem.Lett., 11, 2020
|
|
1RZJ
 
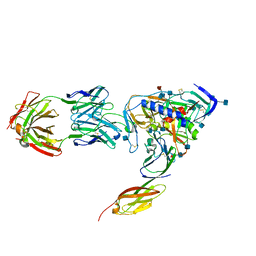 | | HIV-1 HXBC2 GP120 ENVELOPE GLYCOPROTEIN COMPLEXED WITH CD4 AND INDUCED NEUTRALIZING ANTIBODY 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Huang, C.C, Venturi, M, Majeed, S, Moore, M.J, Phogat, S, Zhang, M.-Y, Dimitrov, D.S, Hendrickson, W.A, Robinson, J, Sodroski, J, Wyatt, R, Choe, H, Farzan, M, Kwong, P.D. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6OOT
 
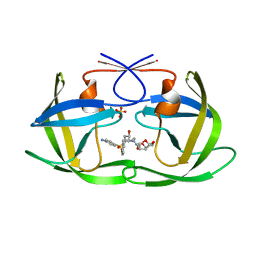 | | HIV-1 Protease NL4-3 L89V, L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
6WNR
 
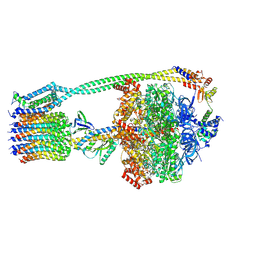 | | E. coli ATP synthase State 3b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
6WYI
 
 | | Crystal structure of EchA19, enoyl-CoA hydratase from Mycobacterium tuberculosis | | Descriptor: | EchA19, enoyl-CoA hydratase, MAGNESIUM ION | | Authors: | Bonds, A.C, Garcia-Diaz, M, Sampson, N.S. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Post-translational Succinylation ofMycobacterium tuberculosisEnoyl-CoA Hydratase EchA19 Slows Catalytic Hydration of Cholesterol Catabolite 3-Oxo-chol-4,22-diene-24-oyl-CoA.
Acs Infect Dis., 6, 2020
|
|
6WPG
 
 | | Structural Basis of Salicylic Acid Perception by Arabidopsis NPR Proteins | | Descriptor: | 2-HYDROXYBENZOIC ACID, Regulatory protein NPR4 | | Authors: | Wang, W, Withers, J, Li, H, Zwack, P.J, Rusnac, D.V, Shi, H, Liu, L, Yan, S, Hinds, T.R, Guttman, M, Dong, X, Zheng, N. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.283 Å) | | Cite: | Structural basis of salicylic acid perception by Arabidopsis NPR proteins.
Nature, 586, 2020
|
|
1XWG
 
 | | Human GST A1-1 T68E mutant | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Novotny, M, Grehn, L, Olin, B, Madsen, D, Wahlberg, M, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2004-11-01 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1SQP
 
 | | Crystal Structure Analysis of Bovine Bc1 with Myxothiazol | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
1XZ9
 
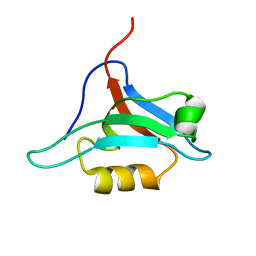 | | Structure of AF-6 PDZ domain | | Descriptor: | Afadin | | Authors: | Joshi, M, Boisguerin, P, Leitner, D, Volkmer-Engert, R, Moelling, K, Schade, M, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2004-11-12 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
6OVF
 
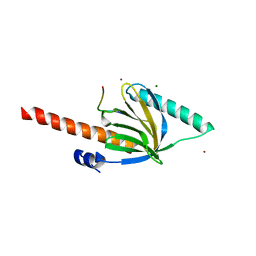 | |
3MRD
 
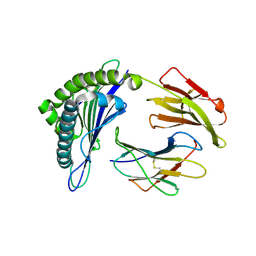 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCMV pp65-495-503 nonapeptide V6G variant | | Descriptor: | 9-meric peptide from Tegument protein pp65, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Reiser, J.-B, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
4R32
 
 | | Crystal Structure Analysis of Pyk2 and Paxillin LD motifs | | Descriptor: | Paxillin, Protein-tyrosine kinase 2-beta | | Authors: | Vanarotti, M, Miller, D.J, Guibao, C.D, Nourse, A, Zheng, J.J. | | Deposit date: | 2014-08-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.505 Å) | | Cite: | Structural and Mechanistic Insights into the Interaction between Pyk2 and Paxillin LD Motifs.
J.Mol.Biol., 426, 2014
|
|
6WQ3
 
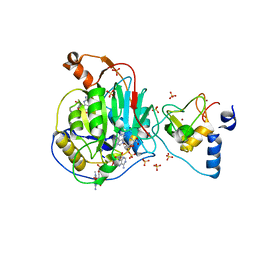 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-adenosyl-L-homocysteine. | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6U12
 
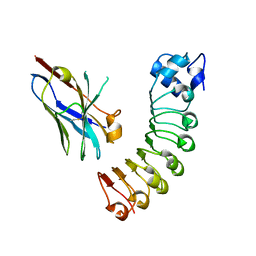 | | VHH R303 C33A/C102A in complex withthe LRR domain of InlB | | Descriptor: | InlB, VHH R303 C33A/C102A mutant | | Authors: | Mendoza, M.N, Jian, M, Toride King, M, Brooks, C.L. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Role of a noncanonical disulfide bond in the stability, affinity, and flexibility of a VHH specific for the Listeria virulence factor InlB.
Protein Sci., 29, 2020
|
|
6WRZ
 
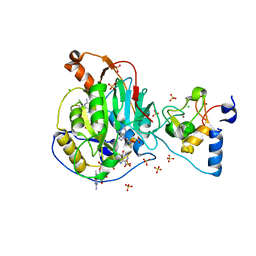 | | Crystal Structure of Nsp16-Nsp10 Heterodimer from SARS-CoV-2 with 7-methyl-GpppA and S-adenosyl-L-homocysteine in the Active Site and Sulfates in the mRNA Binding Groove. | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
6OGS
 
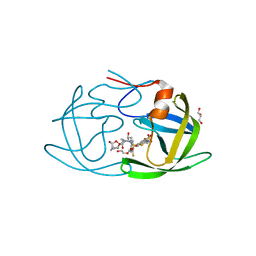 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
1BUD
 
 | | ACUTOLYSIN A FROM SNAKE VENOM OF AGKISTRODON ACUTUS AT PH 5.0 | | Descriptor: | CALCIUM ION, PROTEIN (ACUTOLYSIN A), ZINC ION | | Authors: | Gong, W, Zhu, X, Liu, S, Teng, M, Niu, L. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus.
J.Mol.Biol., 283, 1998
|
|
1XPA
 
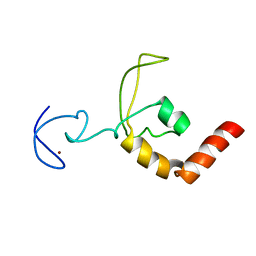 | | SOLUTION STRUCTURE OF THE DNA-AND RPA-BINDING DOMAIN OF THE HUMAN REPAIR FACTOR XPA, NMR, 1 STRUCTURE | | Descriptor: | XPA, ZINC ION | | Authors: | Ikegami, T, Kuraoka, I, Saijo, M, Kodo, N, Kyogoku, Y, Morikawa, K, Tanaka, K, Shirakawa, M. | | Deposit date: | 1998-07-06 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA.
Nat.Struct.Biol., 5, 1998
|
|
8AAN
 
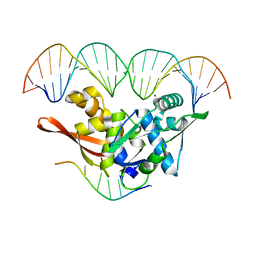 | | The nucleoprotein complex of Rep protein with iteron containing dsDNA and DUE ssDNA. | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*T)-3'), ... | | Authors: | Nowacka, M, Wegrzyn, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Rep protein accommodates together dsDNA and ssDNA which enables a loop-back mechanism to plasmid DNA replication initiation.
Nucleic Acids Res., 51, 2023
|
|
6WTT
 
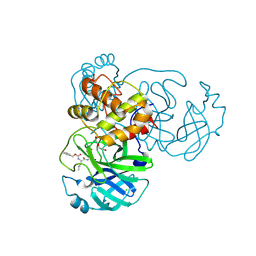 | | Crystals Structure of the SARS-CoV-2 (COVID-19) main protease with inhibitor GC-376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Sacco, M, Ma, C, Chen, Y, Wang, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease.
Cell Res., 30, 2020
|
|
