5V75
 
 | |
5V7V
 
 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
5VBU
 
 | |
5VCN
 
 | | THE CRYSTAL STRUCTURE OF DER P 1 ALLERGEN COMPLEXED WITH FAB FRAGMENT OF MAB 5H8 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Osinski, T, Majorek, K.A, Pomes, A, Offermann, L.R, Osinski, S, Glesner, J, Vailes, L.D, Chapman, M.D, Minor, W, Chruszcz, M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Der p 1-Antibody Complexes and Comparison with Complexes of Proteins or Peptides with Monoclonal Antibodies.
J. Immunol., 195, 2015
|
|
3ACS
 
 | |
5UJV
 
 | | Crystal structure of FePYR1 in complex with abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, PYR1 | | Authors: | Ren, Z, Wang, Z, Zhou, X.E, Hong, Y, Cao, M, Chan, Z, Liu, X, Shi, H, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1.
Sci Rep, 7, 2017
|
|
3VKK
 
 | | Crystal Structure Of The Covalent Intermediate Of Human Cytosolic Beta-Glucosidase-mannose complex | | Descriptor: | CHLORIDE ION, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Okino, N, Ito, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition mechanism of human cytosolic beta-glucosidase by monnoside
To be Published
|
|
5VAI
 
 | | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein | | Descriptor: | Glucagon-like peptide 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y, Sun, B, Feng, D, Hu, H, Chu, M, Qu, Q, Tarrasch, J.T, Li, S, Kobilka, T.S, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2017-03-27 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Nature, 546, 2017
|
|
5UL7
 
 | |
1SRY
 
 | |
3VHJ
 
 | |
5UMZ
 
 | |
2PTF
 
 | | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Uncharacterized protein MTH_863 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of protein MTH_863 from Methanobacterium thermoautotrophicum bound to FMN.
To be Published
|
|
5UMG
 
 | | Crystal structure of dihydropteroate synthase from Klebsiella pneumoniae subsp. | | Descriptor: | Dihydropteroate synthase, GLYCEROL | | Authors: | Chang, C, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-27 | | Release date: | 2017-02-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of dihydropteroate synthase from Klebsiella pneumoniae subsp.
To Be Published
|
|
1JAO
 
 | | COMPLEX OF 3-MERCAPTO-2-BENZYLPROPANOYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2S)-2-benzyl-3-sulfanylpropanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
1JAN
 
 | | COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (PHE79 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (PHE79 FORM), PRO-LEU-GLY-HYDROXYLAMINE INHIBITOR, ... | | Authors: | Reinemer, P, Grams, F, Huber, R, Kleine, T, Schnierer, S, Pieper, M, Tschesche, H, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural implications for the role of the N terminus in the 'superactivation' of collagenases. A crystallographic study.
FEBS Lett., 338, 1994
|
|
5UQS
 
 | |
3AF2
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with AMPPCP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Pantothenate kinase | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3AFJ
 
 | |
1THX
 
 | | THIOREDOXIN-2 | | Descriptor: | THIOREDOXIN | | Authors: | Saarinen, M, Gleason, F.K, Eklund, H. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of thioredoxin-2 from Anabaena.
Structure, 3, 1995
|
|
2N8Y
 
 | | Holo form of Calmodulin-Like Domain of Human Non-Muscle alpha-actinin 1 | | Descriptor: | Alpha-actinin-1, CALCIUM ION | | Authors: | Drmota Prebil, S, Slapsak, U, de Almeida Ribeiro, E, Pavsic, M, Ilc, G, Zielinska, K, Hartl, M, Backman, L, Plavec, J, Lenarcic, B, Djinovic-Carugo, K. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of calmodulin-like domain of human non-muscle alpha-actinin-1.
Sci Rep, 6, 2016
|
|
2N5M
 
 | | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8 | | Descriptor: | Protein KHNYN | | Authors: | Santonico, E, Nepravishta, R, Mattioni, A, Valentini, E, Mandaliti, W, Procopio, R, Iannuccelli, M, Castagnoli, L, Polo, S, Paci, M, Cesareni, G. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unveiling the structural determinants of KIAA0323 binding preference for NEDD8.
To be Published
|
|
1TMR
 
 | | THE STRUCTURE OF A 19 RESIDUE FRAGMENT FROM THE C-LOOP OF THE FOURTH EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF THROMBOMODULIN | | Descriptor: | THROMBOMODULIN PRECURSOR | | Authors: | Adler, M, Seto, M, Nitecki, D, Light, D.R, Morser, J. | | Deposit date: | 1994-05-20 | | Release date: | 1995-06-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The structure of a 19-residue fragment from the C-loop of the fourth epidermal growth factor-like domain of thrombomodulin.
J.Biol.Chem., 270, 1995
|
|
5UK6
 
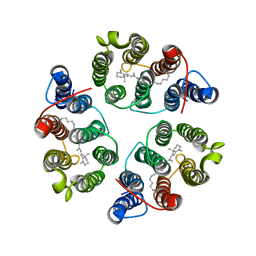 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy and DEER | | Descriptor: | Bacteriorhodopsin | | Authors: | Milikisiyants, S, Wang, S, Munro, R.A, Donohue, M, Ward, M.E, Brown, L.S, Smirnova, T.I, Ladizhansky, V, Smirnov, A.I. | | Deposit date: | 2017-01-20 | | Release date: | 2017-05-31 | | Last modified: | 2020-01-08 | | Method: | SOLID-STATE NMR | | Cite: | Oligomeric Structure of Anabaena Sensory Rhodopsin in a Lipid Bilayer Environment by Combining Solid-State NMR and Long-range DEER Constraints.
J. Mol. Biol., 429, 2017
|
|
5UMM
 
 | |
