1PI8
 
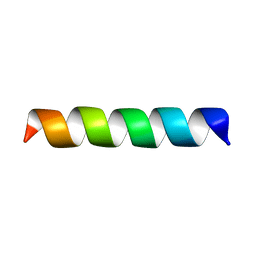 | | Structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1 | | 分子名称: | VPU protein | | 著者 | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | 登録日 | 2003-05-29 | | 公開日 | 2003-11-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Three-dimensional structure of the channel-forming trans-membrane domain of virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
4OMW
 
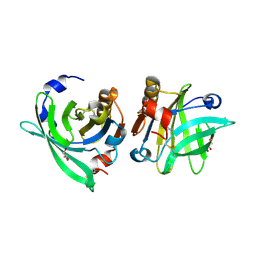 | | Crystal structure of goat beta-lactoglobulin (orthorhombic form) | | 分子名称: | Beta-lactoglobulin, GLYCEROL, SULFATE ION, ... | | 著者 | Loch, J.I, Swiatek, S, Czub, M, Ludwikowska, M, Lewinski, K. | | 登録日 | 2014-01-27 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformational variability of goat beta-lactoglobulin: Crystallographic and thermodynamic studies.
Int.J.Biol.Macromol., 72C, 2014
|
|
1TG7
 
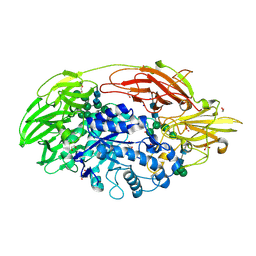 | | Native structure of beta-galactosidase from Penicillium sp. | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | 登録日 | 2004-05-28 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
2CQL
 
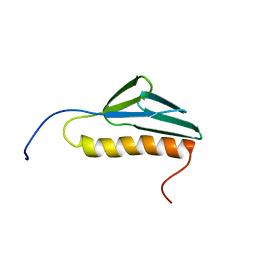 | | Solution structure of the N-terminal domain of human ribosomal protein L9 | | 分子名称: | 60S ribosomal protein L9 | | 著者 | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-20 | | 公開日 | 2005-11-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal domain of human ribosomal protein L9
To be Published
|
|
4OQ2
 
 | | 5hmC specific restriction endonuclease PvuRTs1I | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Restriction endonuclease PvuRts1 I | | 著者 | Kazrani, A.A, Kowalska, M, Czapinska, H, Bochtler, M. | | 登録日 | 2014-02-07 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of the 5hmC specific endonuclease PvuRts1I.
Nucleic Acids Res., 42, 2014
|
|
2CZU
 
 | | lipocalin-type prostaglandin D synthase | | 分子名称: | Prostaglandin-H2 D-isomerase | | 著者 | Kumasaka, T, Irikura, D, Ago, H, Aritake, K, Yamamoto, M, Inoue, T, Miyano, M, Urade, Y, Hayaishi, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-17 | | 公開日 | 2006-10-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of the catalytic mechanism operating in open-closed conformers of lipocalin type prostaglandin D synthase.
J.Biol.Chem., 284, 2009
|
|
7ERO
 
 | | Crystal structure of D-allulose 3-epimerase with D-allulose from Agrobacterium sp. SUL3 | | 分子名称: | D-psicose, D-tagatose 3-epimerase, MAGNESIUM ION | | 著者 | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | 登録日 | 2021-05-06 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERN
 
 | | Crystal structure of D-allulose 3-epimerase with D-fructose from Agrobacterium sp. SUL3 | | 分子名称: | D-fructose, D-tagatose 3-epimerase, MAGNESIUM ION | | 著者 | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | 登録日 | 2021-05-06 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERM
 
 | | Crystal structure of D-allulose 3-epimerase from Agrobacterium sp. SUL3 | | 分子名称: | D-tagatose 3-epimerase, MAGNESIUM ION, SULFATE ION | | 著者 | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | 登録日 | 2021-05-06 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
2MJ2
 
 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | 分子名称: | Agnoprotein | | 著者 | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | 登録日 | 2013-12-23 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
3HGF
 
 | | Expression, purification, spectroscopical and crystallographical studies of segments of the nucleotide binding domain of the reticulocyte binding protein Py235 of Plasmodium yoelii | | 分子名称: | Rhoptry protein fragment | | 著者 | Gruber, A, Manimekalai, M.S.S, Balakrishna, A.M, Hunke, C, Jeyakanthan, J, Preiser, P.R, Gruber, G. | | 登録日 | 2009-05-13 | | 公開日 | 2010-02-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural determination of functional units of the nucleotide binding domain (NBD94) of the reticulocyte binding protein Py235 of Plasmodium yoelii
Plos One, 5, 2010
|
|
5TJF
 
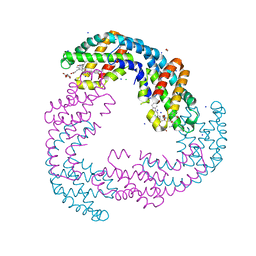 | | The crystal structure of Allophycocyanin from the red algae Gracilaria chilensis | | 分子名称: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, CHLORIDE ION, ... | | 著者 | Figueroa, M, Dagnino, J, Kerff, F, Chartier, P, Bunster, M, Martinez-Oyanedel, J. | | 登録日 | 2016-10-04 | | 公開日 | 2017-05-24 | | 最終更新日 | 2017-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural models of the different trimers present in the core of phycobilisomes from Gracilaria chilensis based on crystal structures and sequences.
PLoS ONE, 12, 2017
|
|
2QO4
 
 | | Crystal structure of zebrafish liver bile acid-binding protein complexed with cholic acid | | 分子名称: | CHOLIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Capaldi, S, Saccomani, G, Perduca, M, Monaco, H.L. | | 登録日 | 2007-07-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Single Amino Acid Mutation in Zebrafish (Danio rerio) Liver Bile Acid-binding Protein Can Change the Stoichiometry of Ligand Binding.
J.Biol.Chem., 282, 2007
|
|
1B5Z
 
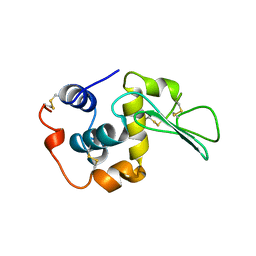 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | 分子名称: | LYSOZYME | | 著者 | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1999-01-11 | | 公開日 | 1999-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
4OXF
 
 | |
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | 登録日 | 2016-09-13 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
2R1R
 
 | |
1B6Y
 
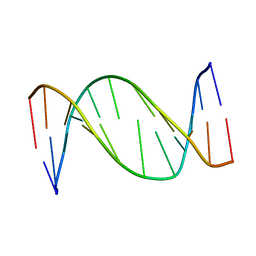 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | 分子名称: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | 著者 | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | 登録日 | 1999-01-19 | | 公開日 | 1999-01-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
2R3X
 
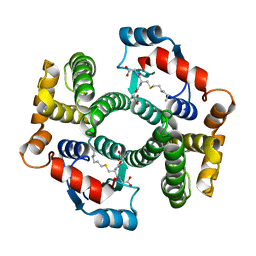 | | Crystal structure of an R15L hGSTA1-1 mutant complexed with S-hexyl-glutathione | | 分子名称: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | 著者 | Burke, J.P.W.G, Kinsley, N, Sayed, M, Sewell, T, Dirr, H.W. | | 登録日 | 2007-08-30 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Arginine 15 stabilizes an S(N)Ar reaction transition state and the binding of anionic ligands at the active site of human glutathione transferase A1-1.
Biophys.Chem., 146, 2010
|
|
4LNY
 
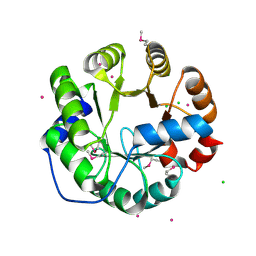 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | 分子名称: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | 著者 | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-07-12 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.929 Å) | | 主引用文献 | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
1BZV
 
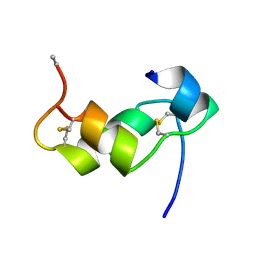 | | [D-ALAB26]-DES(B27-B30)-INSULIN-B26-AMIDE A SUPERPOTENT SINGLE-REPLACEMENT INSULIN ANALOGUE, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | INSULIN | | 著者 | Kurapkat, G, Siedentopf, M, Gattner, H.G, Hagelstein, M, Brandenburg, D, Grotzinger, J, Wollmer, A. | | 登録日 | 1998-11-04 | | 公開日 | 1999-05-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a superpotent B-chain-shortened single-replacement insulin analogue.
Protein Sci., 8, 1999
|
|
4PI8
 
 | | Crystal structure of catalytic mutant E138A of S. Aureus Autolysin E in complex with disaccharide NAG-NAM | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | 著者 | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | 登録日 | 2014-05-08 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
4BUZ
 
 | | SIR2 COMPLEX STRUCTURE MIXTURE OF EX-527 INHIBITOR AND REACTION PRODUCTS OR OF REACTION SUBSTRATES P53 PEPTIDE AND NAD | | 分子名称: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, 1,2-ETHANEDIOL, 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, ... | | 著者 | Weyand, M, Lakshminarasimhan, M, Gertz, M, Steegborn, C. | | 登録日 | 2013-06-24 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5T7M
 
 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-TRP | | 分子名称: | ACETATE ION, Chemotaxis protein, SODIUM ION, ... | | 著者 | Gavira, J.A, Rico-Jimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | 登録日 | 2016-09-05 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5T8F
 
 | | p110delta/p85alpha with taselisib (GDC-0032) | | 分子名称: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Moertl, M, Steinbacher, S, Eigenbrot, C. | | 登録日 | 2016-09-07 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
