8GJN
 
 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | 著者 | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | 登録日 | 2023-03-16 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8G5Z
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state) | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | 著者 | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | 登録日 | 2023-02-14 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8HR3
 
 | | [D-Cys5,Asp7,Val8,D-Lys16]-STp(5-17) | | 分子名称: | DCY-CYS-ASP-VAL-CYS-CYS-ASN-PRO-ALA-CYS-ALA-DLY-CYS | | 著者 | Shimamoto, S, Hidaka, Y, Yoshino, S, Goto, M. | | 登録日 | 2022-12-14 | | 公開日 | 2023-09-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Molecular Basis of Heat-Stable Enterotoxin for Vaccine Development and Cancer Cell Detection.
Molecules, 28, 2023
|
|
6RWX
 
 | |
5LWM
 
 | | Crystal structure of JAK3 in complex with Compound 4 (FM381) | | 分子名称: | 1,2-ETHANEDIOL, 1-phenylurea, 2-cyano-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl)furan-2-yl]-~{N},~{N}-dimethyl-prop-2-enamide, ... | | 著者 | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-18 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
2V9X
 
 | | E138D variant of Escherichia coli dCTP deaminase in complex with dUTP | | 分子名称: | DEOXYCYTIDINE TRIPHOSPHATE DEAMINASE, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Thymark, M, Johansson, E, Larsen, S, Willemoes, M. | | 登録日 | 2007-08-28 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mutational Analysis of the Nucleotide Binding Site of Escherichia Coli Dctp Deaminase.
Arch.Biochem.Biophys., 470, 2008
|
|
7CIG
 
 | | Crystal structure of L-methionine decarboxylase Q64A mutant from Streptomyces sp.590 in complexed with L- methionine methyl ester (geminal diamine form). | | 分子名称: | L-methionine decarboxylase, methyl (2S)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-methylsulfanyl-butanoate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIF
 
 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 (internal aldimine form). | | 分子名称: | L-methionine decarboxylase | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
6S0H
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed doripenem | | 分子名称: | (2~{R},3~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-2,3-dihydro-1~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | 登録日 | 2019-06-14 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6RZ6
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (C2221 space group) | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | 著者 | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | 登録日 | 2019-06-12 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
8GXI
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14c | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The crystal structure of SARS-CoV-2 main protease in complex with 14c
To Be Published
|
|
3MRG
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide | | 分子名称: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, CITRIC ACID, ... | | 著者 | Gras, S, Reiser, J.-B, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | 登録日 | 2010-04-29 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
7AAP
 
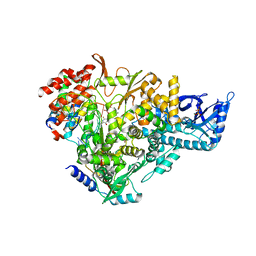 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | 分子名称: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | 著者 | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | 登録日 | 2020-09-04 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3MRR
 
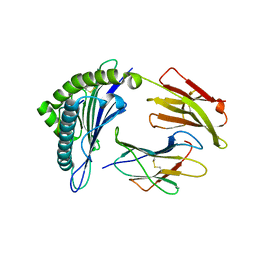 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with Human Prostaglandin Transporter decapeptide | | 分子名称: | 10-meric peptide from Solute carrier organic anion transporter family member 2A1, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | 著者 | Reiser, J.-B, Machillot, P, Chouquet, A, Debeaupuis, E, Echasserieau, K, Legoux, F, Saulquin, X, Bonneville, M, Housset, D. | | 登録日 | 2010-04-29 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
6S3F
 
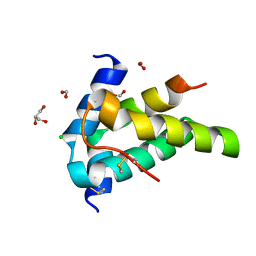 | | Moringa seed protein Mo-CBP3-4 | | 分子名称: | 2S albumin, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Moulin, M, Mossou, E, Mitchell, E.P, Haertlein, M, Forsyth, V.T, Rennie, A.R. | | 登録日 | 2019-06-25 | | 公開日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Towards a molecular understanding of the water purification properties of Moringa seed proteins.
J Colloid Interface Sci, 554, 2019
|
|
8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The crystal structure of SARS-CoV-2 main protease in complex with 14a
To Be Published
|
|
2VBV
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and FMN | | 分子名称: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | 登録日 | 2007-09-16 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
6CNY
 
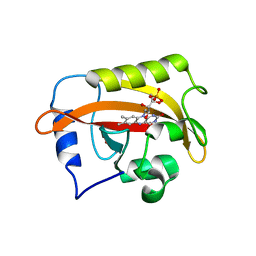 | | 2.3 Angstrom Structure of Phosphodiesterase treated Vivid (complex with FMN) | | 分子名称: | FLAVIN MONONUCLEOTIDE, Vivid PAS protein VVD | | 著者 | Zoltowski, B.D, Shabalin, I.G, Kowiel, M, Porebski, P.J, Crane, B.R, Bilwes, A.M. | | 登録日 | 2018-03-09 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conformational switching in the fungal light sensor Vivid.
Science, 316, 2007
|
|
1JDE
 
 | | K22A mutant of pyruvate, phosphate dikinase | | 分子名称: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | 著者 | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | 登録日 | 2001-06-13 | | 公開日 | 2001-11-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
2UYQ
 
 | | Crystal structure of ML2640c from Mycobacterium leprae in complex with S-adenosylmethionine | | 分子名称: | HYPOTHETICAL PROTEIN ML2640, S-ADENOSYLMETHIONINE | | 著者 | Grana, M, Buschiazzo, A, Wehenkel, A, Haouz, A, Miras, I, Shepard, W, Alzari, P.M. | | 登録日 | 2007-04-11 | | 公開日 | 2007-08-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Crystal Structure of M. Leprae Ml2640C Defines a Large Family of Putative S-Adenosylmethionine- Dependent Methyltransferases in Mycobacteria.
Protein Sci., 16, 2007
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2022-09-20 | | 公開日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | The crystal structure of SARS-CoV-2 main protease in complex with 14b
To Be Published
|
|
6S2W
 
 | | Structure of S. pombe Erh1, a protein important for meiotic mRNA decay in mitosis and meiosis progression. | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Hazra, D, Graille, M. | | 登録日 | 2019-06-22 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Formation of S. pombe Erh1 homodimer mediates gametogenic gene silencing and meiosis progression.
Sci Rep, 10, 2020
|
|
2V5I
 
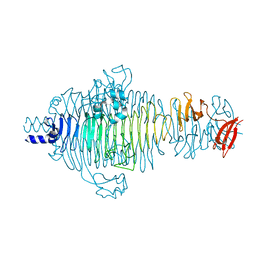 | | Structure of the receptor-binding protein of bacteriophage Det7: a podoviral tailspike in a myovirus | | 分子名称: | SALMONELLA TYPHIMURIUM DB7155 BACTERIOPHAGE DET7 TAILSPIKE, SODIUM ION | | 著者 | Walter, M, Fiedler, C, Grassl, R, Biebl, M, Rachel, R, Hermo-Parrado, X.L, Llamas-Saiz, A.L, Seckler, R, Miller, S, van Raaij, M.J. | | 登録日 | 2007-07-05 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the Receptor-Binding Protein of Bacteriophage Det7: A Podoviral Tail Spike in a Myovirus.
J.Virol., 82, 2008
|
|
6S32
 
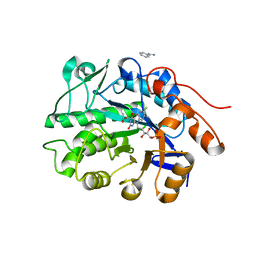 | | Crystal structure of ene-reductase CtOYE from Chroococcidiopsis thermalis. | | 分子名称: | ACETATE ION, BENZAMIDINE, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Robescu, M.R, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | 登録日 | 2019-06-24 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
8GYE
 
 | |
