4QRZ
 
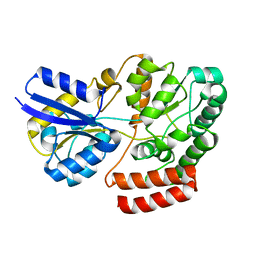 | | Crystal structure of sugar transporter atu4361 from agrobacterium fabrum c58, target efi-510558, with bound maltotriose | | 分子名称: | ABC-TYPE SUGAR TRANSPORTER, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-07-02 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Crystal Structure of Maltoside Transporter from Agrobacterium Radiobacter, Target Efi-510558
To be Published
|
|
6PIQ
 
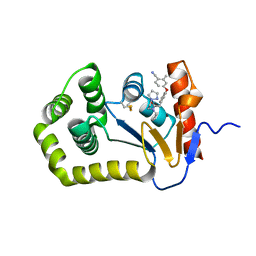 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product G6 (pyrazole 9) | | 分子名称: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | 著者 | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | 登録日 | 2019-06-26 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
4QT4
 
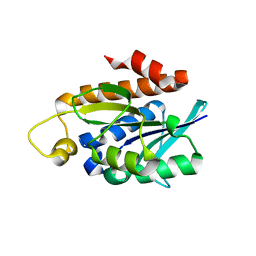 | | Crystal structure of Peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Angstrom resolution shows the Closed Structure of the Substrate Binding Cleft | | 分子名称: | Peptidyl-tRNA hydrolase | | 著者 | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2014-07-07 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
FEBS Open Bio, 4, 2014
|
|
7JW8
 
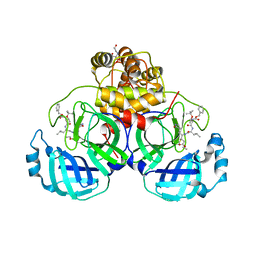 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 in space group P1 | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | 著者 | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | 登録日 | 2020-08-25 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
4RC8
 
 | |
7JSU
 
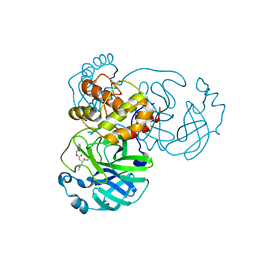 | | Crystal structure of SARS-CoV-2 3CL protease in complex with GC376 | | 分子名称: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, PHOSPHATE ION | | 著者 | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | 登録日 | 2020-08-16 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
8CNX
 
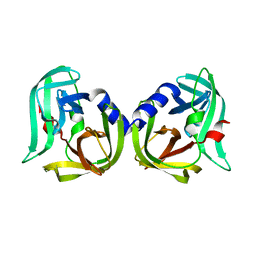 | | Structure of Enterovirus D68 3C protease | | 分子名称: | Protease 3C | | 著者 | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-02-24 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structure of EV D68 3C protease
To Be Published
|
|
4QUX
 
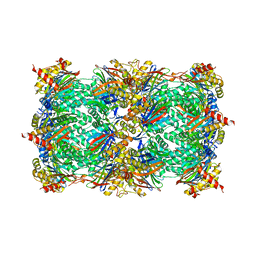 | | yCP beta5-A49T-mutant | | 分子名称: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7JT7
 
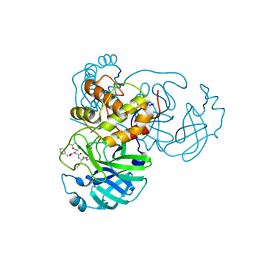 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | 分子名称: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | 著者 | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | 登録日 | 2020-08-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
4RFB
 
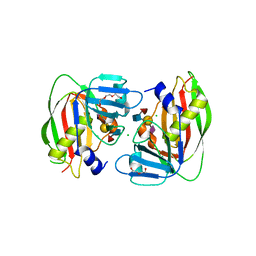 | | 1.93 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with Sialyl-Lewis X. | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-09-25 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | 1.93 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with Sialyl-Lewis X.
TO BE PUBLISHED
|
|
4QVW
 
 | | yCP beta5-A49S-mutant in complex with bortezomib | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-16 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QWM
 
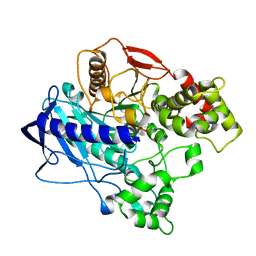 | | KINETIC CRYSTALLOGRAPHY of ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 1.85 MGy | | 分子名称: | DIETHYL HYDROGEN PHOSPHATE, E3 | | 著者 | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | 登録日 | 2014-07-16 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography.
Structure, 24, 2016
|
|
4NP9
 
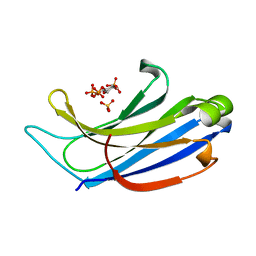 | | Structure of Rabphilin C2A domain bound to IP3 | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rabphilin-3A, SULFATE ION | | 著者 | Guillen, J, Ferrer-Orta, C, Buxaderas, M, Perez-Sanchez, D, Guerrero-Valero, M, Luengo-Gil, G, Pous, J, Guerra, P, Gomez-Fernandez, J.C, Verdaguer, N, Corbalan-Garcia, S. | | 登録日 | 2013-11-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural insights into the Ca2+ and PI(4,5)P2 binding modes of the C2 domains of rabphilin 3A and synaptotagmin 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4QWQ
 
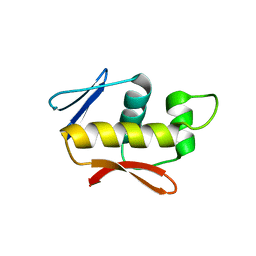 | | Crystal structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus | | 分子名称: | Response regulator SaeR | | 著者 | Fan, X, Zhu, Y, Zhang, X, Teng, M, Li, X. | | 登録日 | 2014-07-17 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | 分子名称: | Protease 3C | | 著者 | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-02-24 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structure of EV D68 3C protease - to be published
To Be Published
|
|
6L9T
 
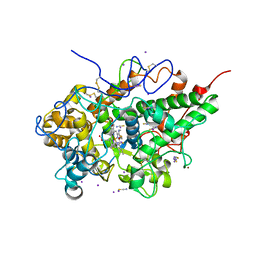 | | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution | | 分子名称: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Singh, P.K, Viswanathan, V, Pandey, N, Singh, A, Sinha, M, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2019-11-11 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution
To Be Published
|
|
4NLI
 
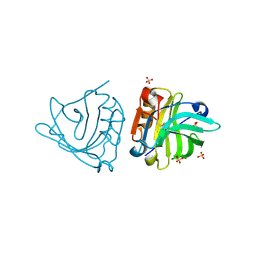 | | Crystal structure of sheep beta-lactoglobulin (space group P3121) | | 分子名称: | Beta-lactoglobulin-1/B, SULFATE ION | | 著者 | Loch, J.I, Molenda, M, Kopec, M, Swiatek, S, Lewinski, K. | | 登録日 | 2013-11-14 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of two crystal forms of sheep beta-lactoglobulin with EF-loop in closed conformation
Biopolymers, 101, 2014
|
|
6PG2
 
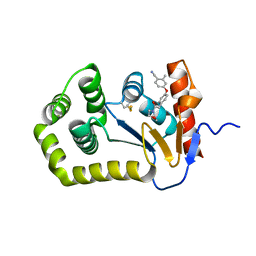 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product H5 (morpholine 8) | | 分子名称: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | 著者 | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | 登録日 | 2019-06-23 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | 分子名称: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | 著者 | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | 登録日 | 2010-08-20 | | 公開日 | 2011-11-23 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3AOU
 
 | | Structure of the Na+ unbound rotor ring modified with N,N f-Dicyclohexylcarbodiimide of the Na+-transporting V-ATPase | | 分子名称: | DICYCLOHEXYLUREA, UNDECYL-MALTOSIDE, V-type sodium ATPase subunit K | | 著者 | Mizutani, K, Yamamoto, M, Yamato, I, Kakinuma, Y, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | 登録日 | 2010-10-06 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4QVP
 
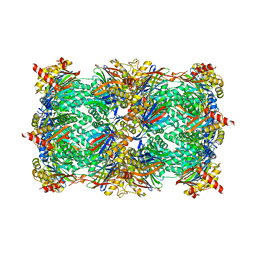 | | yCP beta5-M45T mutant in complex with bortezomib | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-15 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3AI1
 
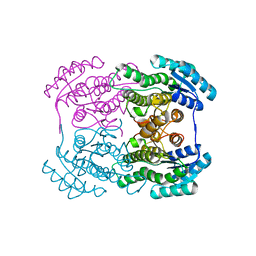 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose reveals the structure bases of its catalytic mechanism and high substrate selectivity | | 分子名称: | NADPH-sorbose reductase | | 著者 | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | 登録日 | 2010-05-06 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
4QWK
 
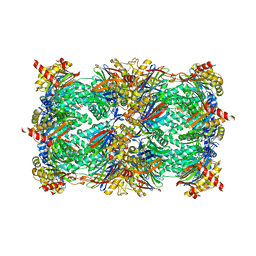 | | yCP beta5-A49T-A50V-double mutant in complex with carfilzomib | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-16 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4R0S
 
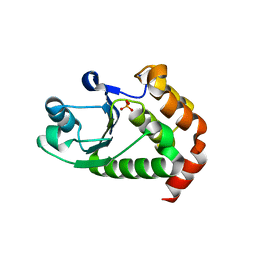 | | Crystal structure of P. aeruginosa TpbA | | 分子名称: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | 著者 | Xu, K, Li, S, Wang, Y, Bartlam, M. | | 登録日 | 2014-08-01 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
4R14
 
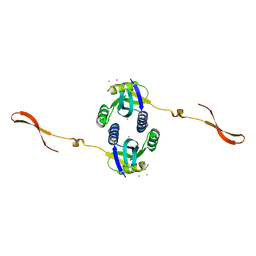 | |
