2ZXM
 
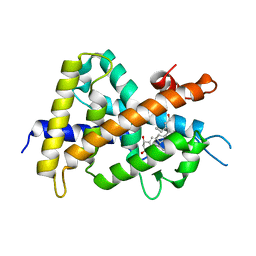 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | 分子名称: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Nakabayashi, M, Ikura, T, Ito, N. | | 登録日 | 2009-01-04 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | 分子名称: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | 著者 | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | 登録日 | 2020-03-20 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
3VGH
 
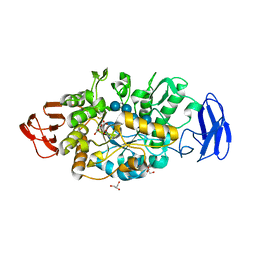 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltotriosyltrehalose | | 分子名称: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | 著者 | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | 登録日 | 2011-08-09 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
5GMU
 
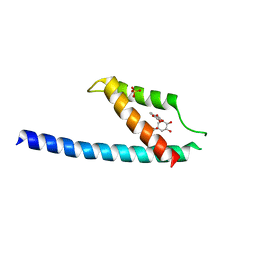 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Chlorogenic acid | | 分子名称: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Protein AroA(G), SULFATE ION | | 著者 | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | 登録日 | 2016-07-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
2MNJ
 
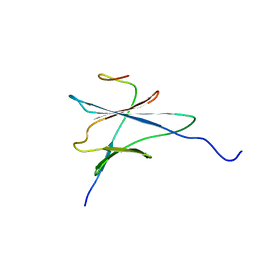 | | NMR solution structure of the yeast Pih1 and Tah1 C-terminal domains complex | | 分子名称: | Protein interacting with Hsp90 1, TPR repeat-containing protein associated with Hsp90 | | 著者 | Quinternet, M, Jacquemin, C, Charpentier, B, Manival, X. | | 登録日 | 2014-04-08 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure/Function Analysis of Protein-Protein Interactions Developed by the Yeast Pih1 Platform Protein and Its Partners in Box C/D snoRNP Assembly.
J.Mol.Biol., 427, 2015
|
|
2ZZ3
 
 | |
1IRG
 
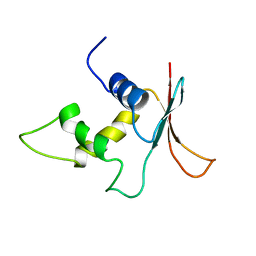 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | INTERFERON REGULATORY FACTOR-2 | | 著者 | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | 登録日 | 1997-11-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
2ZZF
 
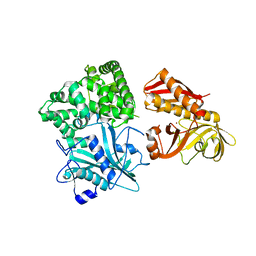 | | Crystal structure of alanyl-tRNA synthetase without oligomerization domain | | 分子名称: | Alanyl-tRNA synthetase, ZINC ION | | 著者 | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | 登録日 | 2009-02-10 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5GNB
 
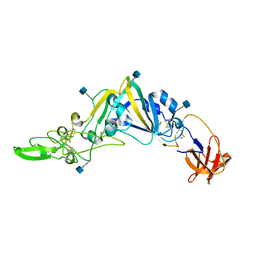 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | 登録日 | 2016-07-20 | | 公開日 | 2017-06-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
2ZYV
 
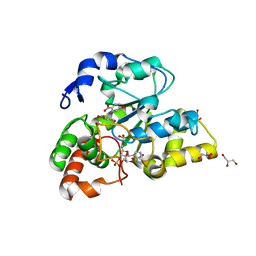 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAPS/PAP and p-nitrophenol | | 分子名称: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, GLYCEROL, P-NITROPHENOL, ... | | 著者 | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | 登録日 | 2009-01-29 | | 公開日 | 2009-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
2ZZ5
 
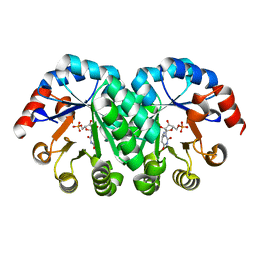 | |
2ZZL
 
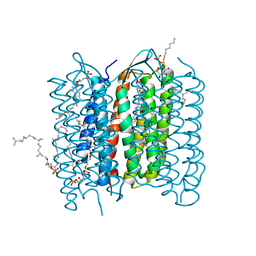 | | Structure of bacteriorhodopsin's M intermediate at pH 7 | | 分子名称: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, ... | | 著者 | Yamamoto, M, Kouyama, T. | | 登録日 | 2009-02-18 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal Structures of Different Substates of Bacteriorhodopsin's M Intermediate at Various pH Levels.
J.Mol.Biol., 2009
|
|
1ITW
 
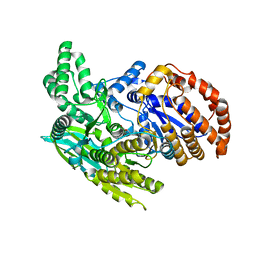 | | Crystal structure of the monomeric isocitrate dehydrogenase in complex with isocitrate and Mn | | 分子名称: | ISOCITRIC ACID, Isocitrate dehydrogenase, MANGANESE (II) ION | | 著者 | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | 登録日 | 2002-02-12 | | 公開日 | 2002-12-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of the Monomeric Isocitrate Dehydrogenase: Evidence of a Protein Monomerization by a Domain Duplication
Structure, 10, 2002
|
|
5GOG
 
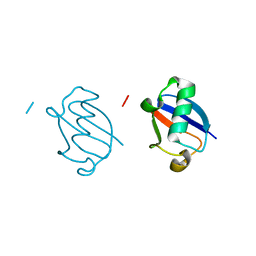 | | Lys29-linked di-ubiquitin | | 分子名称: | D-ubiquitin, Ubiquitin | | 著者 | Gao, S, Pan, M, Zheng, Y. | | 登録日 | 2016-07-27 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.977 Å) | | 主引用文献 | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
5GOK
 
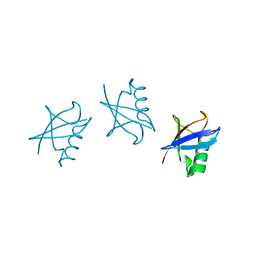 | | K11/K63-branched tri-Ubiquitin | | 分子名称: | D-ubiquitin, Ubiquitin | | 著者 | Gao, S, Pan, M, Zheng, Y, Liu, L. | | 登録日 | 2016-07-27 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
6YPH
 
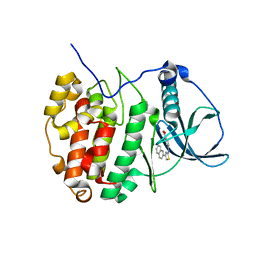 | |
6YPN
 
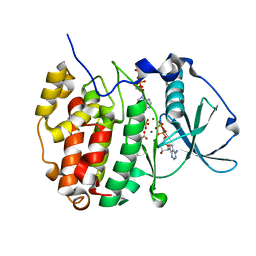 | |
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-06-06 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.91 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
6Y88
 
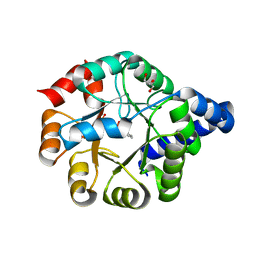 | |
5K65
 
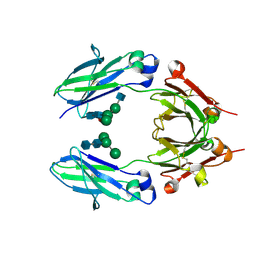 | | Crystal structure of VEGF binding IgG1-Fc (Fcab CT6) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-05-24 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
5GOV
 
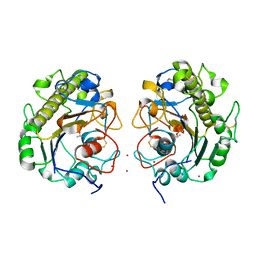 | | Crystal Structure of MCR-1, a phosphoethanolamine transferase, extracellular domain | | 分子名称: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | 著者 | Hu, M, Guo, J, Chen, S, Hao, Q. | | 登録日 | 2016-07-29 | | 公開日 | 2016-12-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal Structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for Colistin Resistance.
Sci Rep, 6, 2016
|
|
6YPS
 
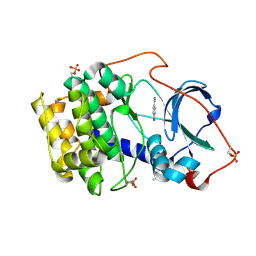 | | Crystal structure of the cAMP-dependent protein kinase A in complex with 4-hydroxybenzamidine | | 分子名称: | 4-oxidanylbenzenecarboximidamide, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | 登録日 | 2020-04-16 | | 公開日 | 2020-12-09 | | 最終更新日 | 2020-12-30 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7SF2
 
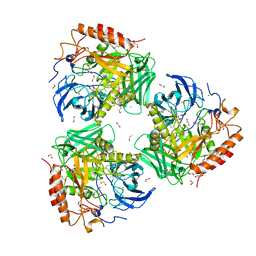 | | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | 著者 | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2021-10-02 | | 公開日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of Beta-Galactosidase from Bacteroides cellulosilyticus
To Be Published
|
|
1J7Y
 
 | | Crystal structure of partially ligated mutant of HbA | | 分子名称: | CARBON MONOXIDE, Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | 登録日 | 2001-05-19 | | 公開日 | 2002-02-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
6EXT
 
 | |
