2RDL
 
 | | Hamster Chymase 2 | | Descriptor: | Chymase 2, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION | | Authors: | Spurlino, J, Abad, M, Kervinen, J. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-30 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for elastolytic substrate specificity in rodent alpha-chymases.
J.Biol.Chem., 283, 2008
|
|
5W6S
 
 | | Crystal structure of Bacteriophage CBA120 tailspike protein 2 enzymatically active domain (TSP2dN, orf211) complex with Escherichia Coli O157-antigen | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Plattner, M, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2017-06-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120.
J.Mol.Biol., 431, 2019
|
|
7RHC
 
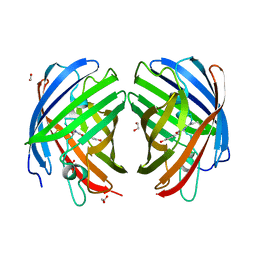 | | A new fluorescent protein darkmRuby at pH 9.0 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
7RHD
 
 | | darkmRuby M94T/F96Y mutant at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby M94T/F96Y mutant | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
6X8Z
 
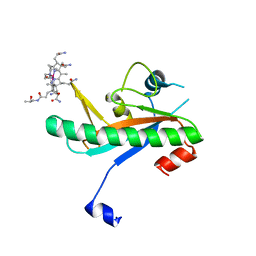 | | Crystal structure of N-truncated human B12 chaperone CblD(C262S)-thiolato-cob(III)alamin complex (108-296) | | Descriptor: | COBALAMIN, Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Mascarenhas, R, Li, Z, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interprotein Co-S Coordination Complex in the B 12 -Trafficking Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
1HAK
 
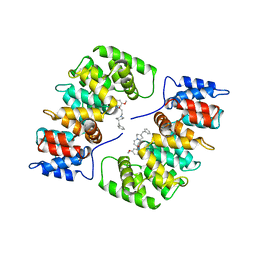 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN PLACENTAL ANNEXIN V COMPLEXED WITH K-201 AS A CALCIUM CHANNEL ACTIVITY INHIBITOR | | Descriptor: | 4-[3-{1-(4-BENZYL)PIPERODINYL}PROPIONYL]-7-METHOXY-2,3,4,5-TERTRAHYDRO-1,4-BENZOTHIAZEPINE, ANNEXIN V | | Authors: | Ago, H, Inagaki, E, Miyano, M. | | Deposit date: | 1997-12-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of annexin V with its ligand K-201 as a calcium channel activity inhibitor.
J.Mol.Biol., 274, 1997
|
|
7R0T
 
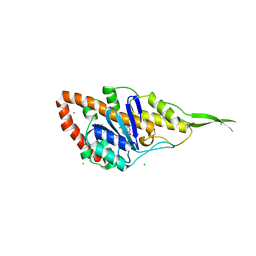 | | Crystal structure of exonuclease ExnV1 | | Descriptor: | CHLORIDE ION, Exonuclease ExnV1, MAGNESIUM ION, ... | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Jasilionis, A, Linares-Pasten, J.A, Wang, L, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R0K
 
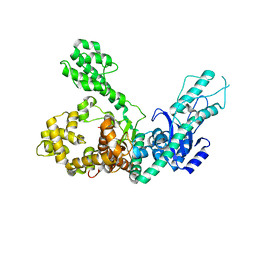 | | Crystal structure of Polymerase I from phage G20c | | Descriptor: | DNA polymerase I | | Authors: | Welin, M, Svensson, A, Hakansson, M, Al-Karadaghi, S, Linares-Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Ahlqvist, J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of DNA polymerase I from Thermus phage G20c.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6CP7
 
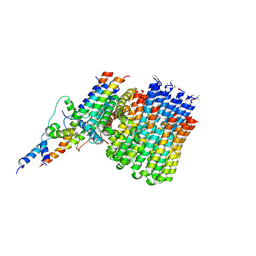 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc generated from masked refinement. | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Srivastava, A.P, Luo, M, Symersky, J, Liao, M.F, Mueller, D.M. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane.
Science, 360, 2018
|
|
6GCA
 
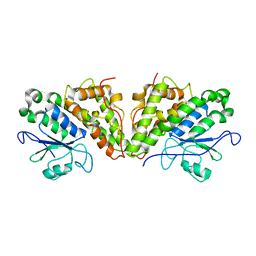 | |
6SRX
 
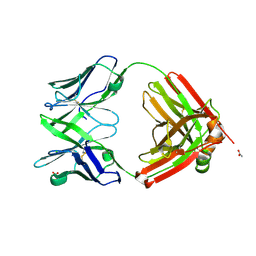 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
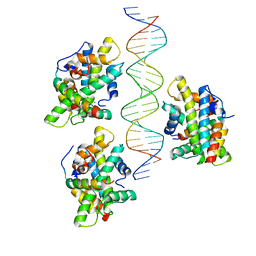
6Y39
 
 | |
5DVH
 
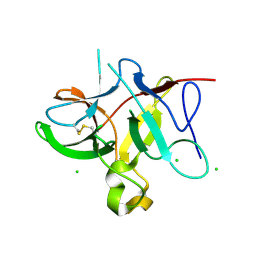 | |
5OJQ
 
 | | The modeled structure of of wild type extended type VI secretion system sheath/tube complex in vibrio cholerae based on cryo-EM reconstruction of the non-contractile sheath/tube complex | | Descriptor: | Haemolysin co-regulated protein, Type VI secretion protein, VipA | | Authors: | Wang, J, Brackmann, M, Castano-Diez, D, Kudryashev, M, Goldie, K, Maier, T, Stahlberg, H, Basler, M. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the extended type VI secretion system sheath-tube complex.
Nat Microbiol, 2, 2017
|
|
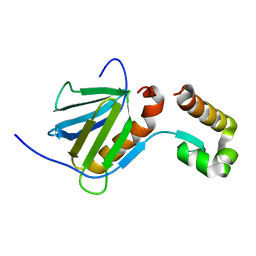
2RNR
 
 | |
6XZO
 
 | | Crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((4-bromo-2-hydroxybenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(4-bromanyl-2-oxidanyl-phenyl)methylamino]ethyl]-3-(4-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6SS6
 
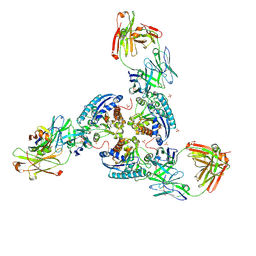 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | Arginase-2, mitochondrial, Fab C0020187 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2RQC
 
 | | Solution Structure of RNA-binding domain 3 of CUGBP1 in complex with RNA (UG)3 | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*G)-3', CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 37, 2009
|
|
2RRB
 
 | | Refinement of RNA binding domain in human Tra2 beta protein | | Descriptor: | cDNA FLJ40872 fis, clone TUTER2000283, highly similar to Homo sapiens transformer-2-beta (SFRS10) gene | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
5W6F
 
 | |
7AQY
 
 | |
7AQZ
 
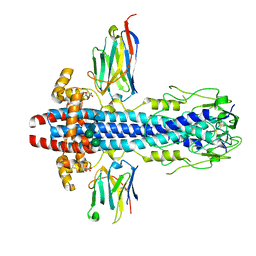 | | Co-Crystal Structure of Variant Surface Glycoprotein VSG2 in complex with Nanobody VSG2(NB14) | | Descriptor: | CITRIC ACID, Nanobody VSG2(NB14), SODIUM ION, ... | | Authors: | Stebbins, C.E, Hempelmann, A, VanStraaten, M. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome.
Cell Rep, 37, 2021
|
|
4IPS
 
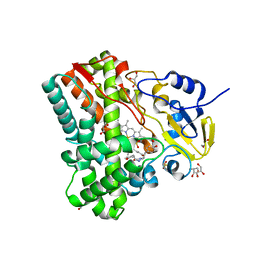 | | Substrate and reaction specificity of Mycobacterium tuberculosis cytochrome P450 CYP121 | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazin-2-one, Cytochrome P450 121, GLYCEROL, ... | | Authors: | Fonvielle, M, LeDu, M.H, Lequin, O, Lecoq, A, Jacquet, M, Thai, R, Dubois, S, Grach, G, Gondry, M, Belin, P. | | Deposit date: | 2013-01-10 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate and Reaction Specificity of Mycobacterium tuberculosis Cytochrome P450 CYP121: INSIGHTS FROM BIOCHEMICAL STUDIES AND CRYSTAL STRUCTURES.
J.Biol.Chem., 288, 2013
|
|
1QB1
 
 | |
6CCK
 
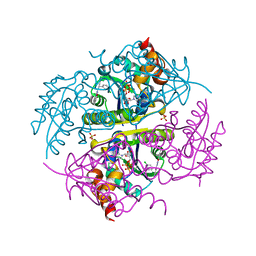 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3-(3-chlorophenyl)-3-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)propanenitrile | | Descriptor: | (3R)-3-(3-chlorophenyl)-3-[(5-methyl-7-oxo-6,7-dihydro[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]propanenitrile, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
