6HVQ
 
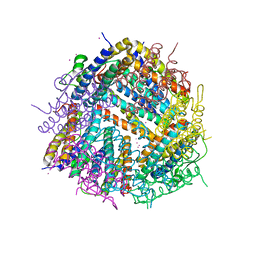 | |
6HXV
 
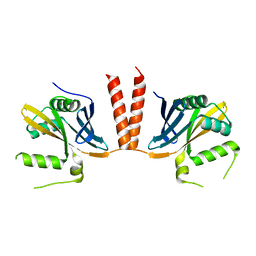 | |
3RCE
 
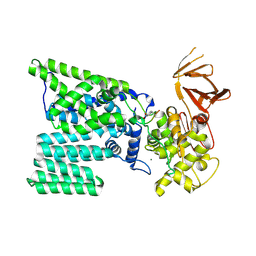 | | Bacterial oligosaccharyltransferase PglB | | Descriptor: | MAGNESIUM ION, Oligosaccharide transferase to N-glycosylate proteins, Substrate Mimic Peptide | | Authors: | Lizak, C, Gerber, S, Numao, S, Aebi, M, Locher, K.P. | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a bacterial oligosaccharyltransferase.
Nature, 474, 2011
|
|
3RKE
 
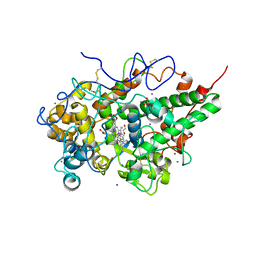 | | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution | | Descriptor: | (4-aminophenyl)-imidazol-1-yl-methanone, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dube, D, Singh, R.P, Sinha, M, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution
TO BE PUBLISHED
|
|
3RKS
 
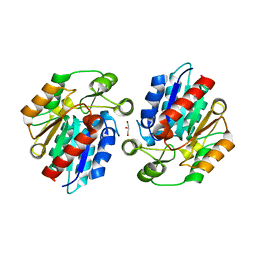 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) K176P mutant | | Descriptor: | GLYCEROL, Hydroxynitrilase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H. | | Deposit date: | 2011-04-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
To be Published
|
|
3RL9
 
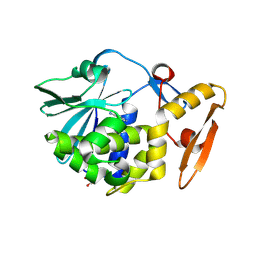 | | Crystal Structure of the complex of type I RIP with the hydrolyzed product of dATP, adenine at 1.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
6HZ1
 
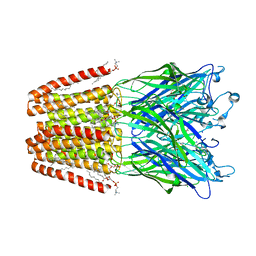 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT E243C | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HBC
 
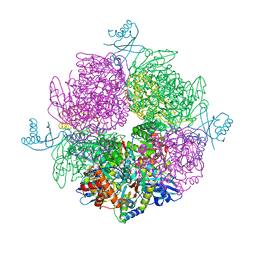 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
3RHI
 
 | | DNA-binding protein HU from Bacillus anthracis | | Descriptor: | DNA-binding protein HU | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | DNA-binding protein HU from Bacillus anthracis.
To be Published
|
|
6HDY
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with S3-HB-AMP | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6I0V
 
 | | Crystal structure of DmTailor in complex with CACAGU RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*AP*CP*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
3RI9
 
 | | Xylanase C from Aspergillus kawachii F131W mutant | | Descriptor: | Endo-1,4-beta-xylanase 3 | | Authors: | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | Deposit date: | 2011-04-13 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
3RO3
 
 | | crystal structure of LGN/mInscuteable complex | | Descriptor: | CHLORIDE ION, ETHANOL, G-protein-signaling modulator 2, ... | | Authors: | Zhu, J, Wen, W, Shang, Y, Wei, Z, Pan, Z, Wang, W, Zhang, M. | | Deposit date: | 2011-04-25 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Mol.Cell, 43, 2011
|
|
6H8J
 
 | | 1.45 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPTO | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-08-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into Urease Inhibition by N-( n-Butyl) Phosphoric Triamide through an Integrated Structural and Kinetic Approach.
J.Agric.Food Chem., 67, 2019
|
|
6H99
 
 | | Crystal structure of anaerobic ergothioneine biosynthesis enzyme from Chlorobium limicola in persulfide form. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Leisinger, F, Burn, R, Meury, M, Lukat, P, Seebeck, F.P. | | Deposit date: | 2018-08-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Basis for Anaerobic Ergothioneine Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
3RQ6
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTA
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Acetyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3R1O
 
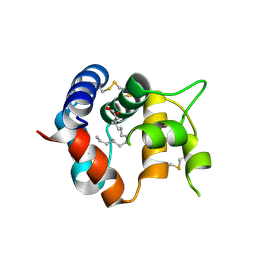 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3RQS
 
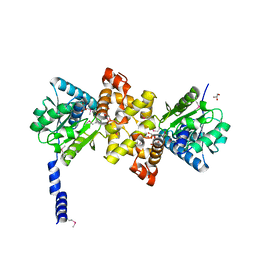 | | Crystal Structure of human L-3- Hydroxyacyl-CoA dehydrogenase (EC1.1.1.35) from mitochondria at the resolution 2.0 A, Northeast Structural Genomics Consortium Target HR487, Mitochondrial Protein Partnership | | Descriptor: | GLYCEROL, Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR487
To be published
|
|
3R5Z
 
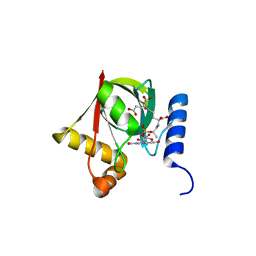 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R2R
 
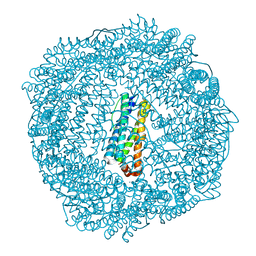 | | 1.65A resolution structure of Iron Soaked FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION, ... | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
5M11
 
 | | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lasica, A.M, Goulas, T, Mizgalska, D, Zhou, X, Ksiazek, M, Madej, M, Guo, Y, Guevara, T, Nowak, M, Potempa, B, Goel, A, Sztukowska, M, Prabhakar, A.T, Bzowska, M, Widziolek, M, Thogersen, I.B, Enghild, J.J, Simonian, M, Kulczyk, A.W, Nguyen, K.-A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis.
Sci Rep, 6, 2016
|
|
3R72
 
 | | Apis mellifera odorant binding protein 5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Odorant binding protein ASP5 | | Authors: | Spinelli, S, Iovinella, I, Lagarde, A, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera odorant binding protein 5
To be Published
|
|
3R5O
 
 | | Crystal structure of the complex of bovine lactoperoxidase with 4-allyl-2-methoxyphenol at 2.6 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, A.K, Singh, R.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-19 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with 4-allyl-2-methoxyphenol at 2.6 A resolution
To be Published
|
|
3R77
 
 | | Crystal structure of the D38A mutant of isochorismatase PhzD from Pseudomonas fluorescens 2-79 in complex with 2-amino-2-desoxyisochorismate ADIC | | Descriptor: | (5S,6S)-6-amino-5-[(1-carboxyethenyl)oxy]cyclohexa-1,3-diene-1-carboxylic acid, CHLORIDE ION, Probable isochorismatase | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
