4QO3
 
 | |
4QO8
 
 | | Lactate Dehydrogenase A in complex with substituted 3-Hydroxy-2-mercaptocyclohex-2-enone compound 104 | | Descriptor: | (5S)-2-[(2-chlorophenyl)sulfanyl]-5-(2,6-dichlorophenyl)-3-hydroxycyclohex-2-en-1-one, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2014-06-19 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Identification of substituted 3-hydroxy-2-mercaptocyclohex-2-enones as potent inhibitors of human lactate dehydrogenase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8R2C
 
 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 598, 2024
|
|
4QAJ
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4QHG
 
 | | Crystal structure of Methanocaldococcus jannaschii dimeric selecase | | Descriptor: | GLYCEROL, Uncharacterized protein MJ1213, ZINC ION | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4QI9
 
 | | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate
To be Published
|
|
8QZK
 
 | | Catalytic core of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) concieved by deep network hallucination: dEngBF4 Hexagonal form | | Descriptor: | ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE | | Authors: | Aghajari, N, Hansen, A.L, Thiesen, F.F, Crehuet, R, Marcos, E, Willemoes, M. | | Deposit date: | 2023-10-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Carving out a Glycoside Hydrolase Active Site for Incorporation into a New Protein Scaffold Using Deep Network Hallucination.
Acs Synth Biol, 13, 2024
|
|
4QG0
 
 | | Crystal structure of the tetrameric dGTP/dUTP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DEOXYURIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
8R4I
 
 | |
3OE5
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, pyridylsulfanyl-containing inhibitor - humanized form | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, N-[(E)-3-[(2R,3S,4R,5S)-3,4-dihydroxy-5-pyridin-4-ylsulfanyl-oxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-nitro-benzamide | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-08-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
3BMP
 
 | |
4QKS
 
 | | Crystal Structure of 6xTrp/PV2: de novo designed beta-trefoil architecture with symmetric primary structure (L22W/L44W/L64W/L85W/L108W/L132W his Primitive Version 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DE NOVO PROTEIN 6XTRP/PV2, SULFATE ION | | Authors: | Longo, L.M, Tenorio, C.A, Blaber, M. | | Deposit date: | 2014-06-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.
Protein Sci., 24, 2015
|
|
6HY2
 
 | |
8RZJ
 
 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor ADG-IF (3,6-anhydro-D-galacto-isofagomine). | | Descriptor: | (1~{R},5~{R},8~{S})-6-oxa-3-azabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4QOD
 
 | | The value crystal structure of apo quinone reductase 2 at 1.35A | | Descriptor: | GLYCEROL, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The value crystal structure of apo quinone reductase 2 at 1.35A
To be Published
|
|
5MC5
 
 | | Crystal Structure of delGlu452 mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, HYDROXIDE ION, ... | | Authors: | Wilk, P, Mueller, U, Dobbek, H, Weiss, M.S. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
4QHT
 
 | | Crystal structure of AAA+/ sigma 54 activator domain of the flagellar regulatory protein FlrC from Vibrio cholerae in ATP analog bound state | | Descriptor: | 1,2-ETHANEDIOL, Flagellar regulatory protein C, MAGNESIUM ION, ... | | Authors: | Dey, S, Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2014-05-29 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.559 Å) | | Cite: | Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC
J.Biol.Chem., 290, 2015
|
|
4QG4
 
 | | Crystal structure of the tetrameric GTP/dATP/ATP-bound SAMHD1 (H210A) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
4QHH
 
 | | Crystal structure of Methanocaldococcus jannaschii tetrameric selecase | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6KLV
 
 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7TDD
 
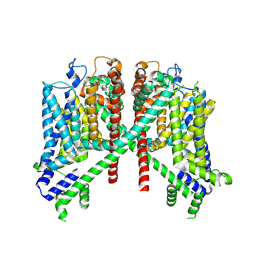 | | AtTPC1 D454N-EDTA state II | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4QMC
 
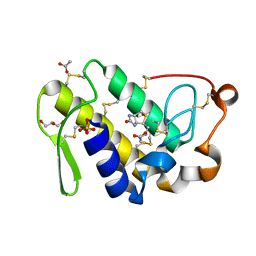 | | Crystal structure of complex formed between phospholipase A2 and Biotin-sulfoxide at 1.09 A Resolution | | Descriptor: | ACETATE ION, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of complex formed between phospholipase A2 and Biotin-sulfoxide at 1.09 A Resolution
To be published
|
|
6KNO
 
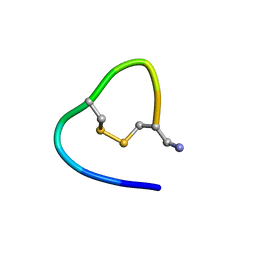 | |
4QO7
 
 | |
6KMY
 
 | | Structure of single disulfide peptide Czon1107-P5A | | Descriptor: | Czon1107-P5A | | Authors: | Sarma, S.P, Madhan Kumar, M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and allosteric activity of a single-disulfide conopeptide fromConus zonatusat human alpha 3 beta 4 and alpha 7 nicotinic acetylcholine receptors.
J.Biol.Chem., 295, 2020
|
|
