6Q4M
 
 | | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, PHOSPHATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation
To Be Published
|
|
6Q56
 
 | | Crystal structure of the B. subtilis M1A22 tRNA methyltransferase TrmK | | Descriptor: | NICKEL (II) ION, tRNA (adenine(22)-N(1))-methyltransferase | | Authors: | Degut, C, Roovers, M, Barraud, P, Brachet, F, Feller, A, Larue, V, Al Refaii, A, Caillet, J, Droogmans, L, Tisne, C. | | Deposit date: | 2018-12-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of B. subtilis m1A22 tRNA methyltransferase TrmK: insights into tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
5ZRS
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl adipate bound state | | Descriptor: | 6-ethoxy-6-oxohexanoic acid, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | Authors: | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
6Q6U
 
 | |
6Q6Z
 
 | |
6Q4R
 
 | | High-resolution crystal structure of ERAP1 with bound phosphinic transition-state analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Neu, M, Rowland, P, Stratikos, E. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of Endoplasmic Reticulum Aminopeptidase 1 with Bound Phosphinic Transition-State Analogue Inhibitor.
Acs Med.Chem.Lett., 10, 2019
|
|
6Q82
 
 | | Crystal structure of the biportin Pdr6 in complex with RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin beta-like protein KAP122, ... | | Authors: | Aksu, M, Vera-Rodriguez, A, Trakhanov, S, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
8QLH
 
 | |
8QLK
 
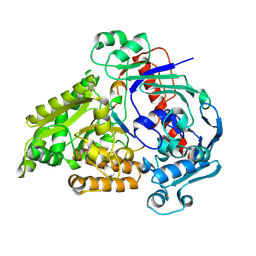 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 2 | | Descriptor: | ALA-ILE-GLN-SER-GLU-LYS-ALA-ARG-LYS-HIS-ASN, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
5ZT6
 
 | |
8QQ5
 
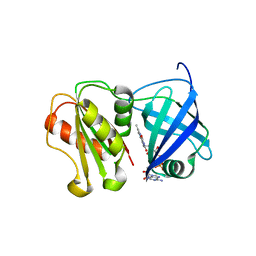 | | Structure of WT SpNox DH domain: a bacterial NADPH oxidase. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Thepaut, M, Petit-Hartlein, I, Vermot, A, Humm, A.S, Dupeux, F, Marquez, J.A, Smith, S, Fieschi, F. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and enzymatic study of a bacterial NADPH oxidase highlight the activation mechanism of eukaryotic NOX.
Elife, 13, 2024
|
|
8QLJ
 
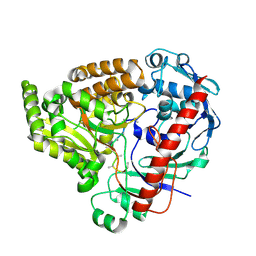 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with an unknown peptide | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
6Q6C
 
 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | 1,2-ETHANEDIOL, Kunitz-type conkunitzin-S1, NITRATE ION, ... | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8QLC
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AliD in open conformation | | Descriptor: | AliD, MAGNESIUM ION | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QPJ
 
 | |
6QGA
 
 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
8QLM
 
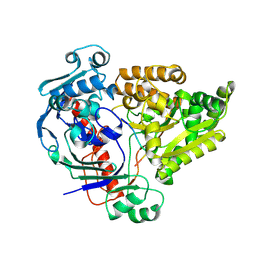 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 3 | | Descriptor: | Oligopeptide-binding protein AliB, PRO-ILE-VAL-GLY-GLY-HIS-GLU-GLY-ALA-GLY-VAL | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QPL
 
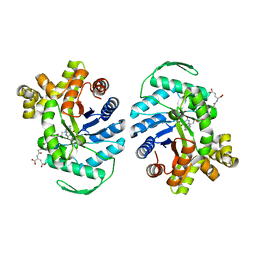 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QQ8
 
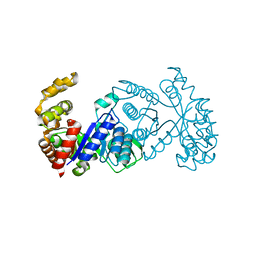 | | Crystal Structure of F420-dependent Methylene-Tetrahydromethanopterin Reductase Mutant E6Q from Methanocaldococcus Jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
1AC9
 
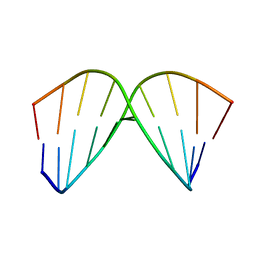 | | SOLUTION STRUCTURE OF A DNA DECAMER CONTAINING THE ANTIVIRAL DRUG GANCICLOVIR: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS, AND FULL RELAXATION REFINEMENT, 6 STRUCTURES | | Descriptor: | DNA | | Authors: | Foti, M, Marshalko, S, Schurter, E, Kumar, S, Beardsley, G.P, Schweitzer, B.I. | | Deposit date: | 1997-02-17 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA decamer containing the antiviral drug ganciclovir: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 36, 1997
|
|
6QD4
 
 | | MloK1 model from single particle analysis of 2D crystals, class 8 (intermediate conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
1A4P
 
 | | P11 (S100A10), LIGAND OF ANNEXIN II | | Descriptor: | S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
6QBP
 
 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203D | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Ramos, N, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203D
To Be Published
|
|
6QEN
 
 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC240 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-[[1-[(4-bromophenyl)methyl]-1,2,3-triazol-4-yl]methylamino]-2-methyl-1-oxidanylidene-propane-2-sulfonic acid, Chymotrypsin-like elastase family member 1, ... | | Authors: | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | Deposit date: | 2019-01-08 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.195 Å) | | Cite: | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6AAO
 
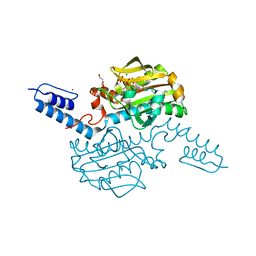 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TCO*Lys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
