8QI9
 
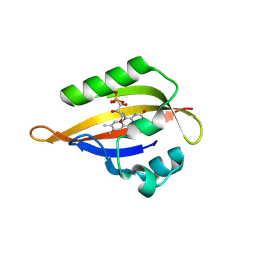 | | CrPhotLOV1 dark state structure determined by serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIK
 
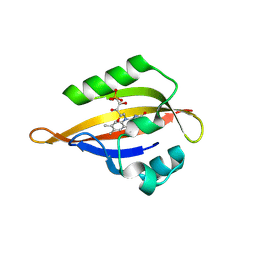 | | CrPhotLOV1 light state structure 32.5 ms (30-35 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIT
 
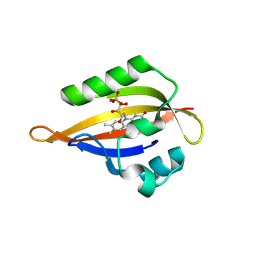 | | CrPhotLOV1 light state structure 77.5 ms (75-80 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QLL
 
 | |
5Y09
 
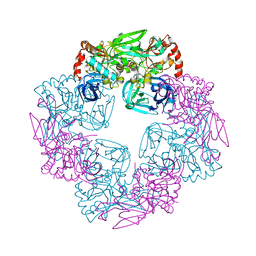 | |
6P9N
 
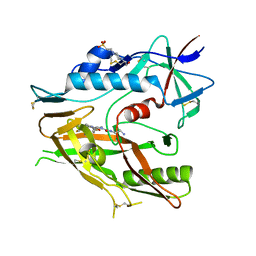 | | CRYSTAL STRUCTURE OF HIV-1 LM/HT CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210. | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-06-10 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
8QUE
 
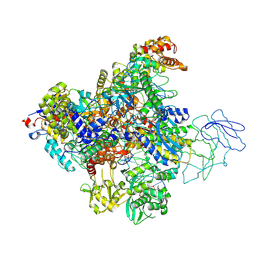 | | Structure of the Bacteriophage PhiKZ non-virion RNA Polymerase bound to DNA and RNA | | Descriptor: | DNA, DNA-directed RNA polymerase, PHIKZ055, ... | | Authors: | de Martin Garrido, N, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2023-10-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Bacteriophage PhiKZ Non-virion RNA Polymerase Transcribing from its Promoter p119L.
J.Mol.Biol., 436, 2024
|
|
6PJC
 
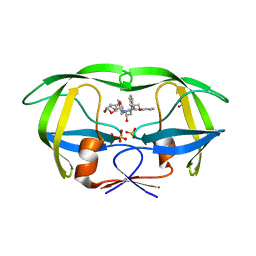 | | HIV-1 Protease NL4-3 WT in Complex with LR4-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-5-{[(2,6-dimethylphenoxy)acetyl]amino}-4-hydroxy-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.965 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8R1O
 
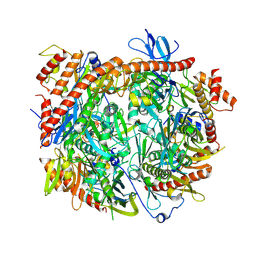 | | Structure of C. thermophilum RNA exosome core | | Descriptor: | Exoribonuclease phosphorolytic domain-containing protein, Exoribonuclease-like protein, Exosome complex component MTR3, ... | | Authors: | Lazzaretti, D, Liebau, J, Pilsl, M, Sprangers, R. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Beyond static structures: quantitative dynamics in the eukaryotic RNA exosome complex
Biorxiv, 2024
|
|
5Y4E
 
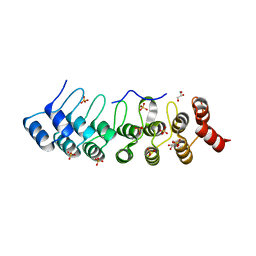 | | Crystal Structure of AnkB Ankyrin Repeats R8-14 in complex with autoinhibition segment AI-b | | Descriptor: | Ankyrin-2,Ankyrin-2, GLYCEROL, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
6PNX
 
 | |
5Y8O
 
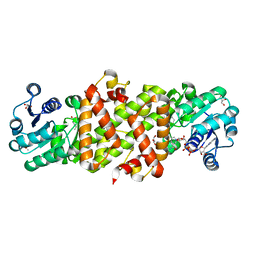 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD + 3-Hydroxy propionate (3-HP) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, 3-HYDROXY-PROPANOIC ACID, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
6PF5
 
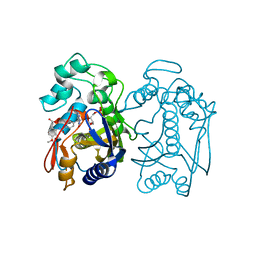 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-methoxyphenyl)acetic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-methoxyphenyl]acetic acid | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6PFF
 
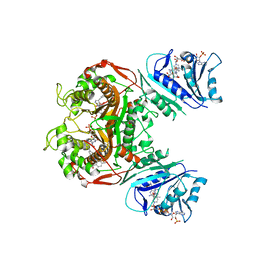 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-cyanobenzoic acid. | | Descriptor: | 2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-cyanobenzoic acid, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
5Y9B
 
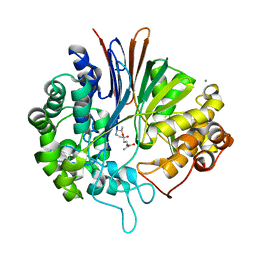 | | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with DON | | Descriptor: | 1,2-ETHANEDIOL, 6-DIAZENYL-5-OXO-L-NORLEUCINE, CALCIUM ION, ... | | Authors: | Goel, M, Kumari, S, Pal, R, Gupta, R. | | Deposit date: | 2017-08-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine
To Be Published
|
|
5XXP
 
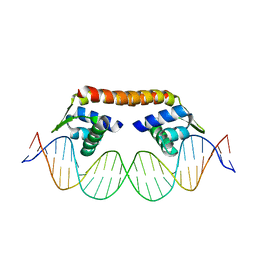 | | Crystal structure of CbnR_DBD-DNA complex | | Descriptor: | DNA (25-MER), LysR-type regulatory protein | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the DNA-binding domain of the LysR-type transcriptional regulator CbnR in complex with a DNA fragment of the recognition-binding site in the promoter region
FEBS J., 285, 2018
|
|
5Y8I
 
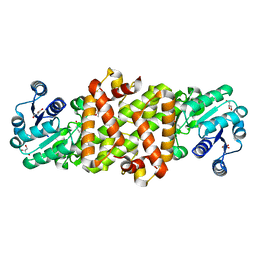 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + (S)-3-hydroxyisobutyrate (S-HIBA) | | Descriptor: | (2S)-2-methyl-3-oxidanyl-propanoic acid, (2~{S})-2-methylpentanedioic acid, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
6PK9
 
 | |
5YE2
 
 | |
8QVM
 
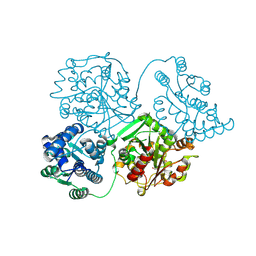 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, TRIETHYLENE GLYCOL | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 2024
|
|
5YF4
 
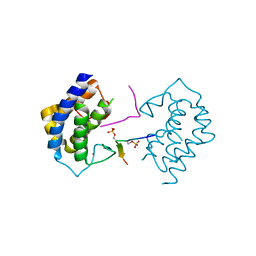 | | A kinase complex MST4-MOB4 | | Descriptor: | MOB-like protein phocein, Peptide from Serine/threonine-protein kinase 26, ZINC ION | | Authors: | Chen, M, Zhou, Z.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer.
J. Biol. Chem., 293, 2018
|
|
8QWG
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, TRIETHYLENE GLYCOL | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 2024
|
|
8QVK
 
 | | Comparison of room-temperature and cryogenic structures of soluble Epoxide Hydrolase with ligands bound. | | Descriptor: | Bifunctional epoxide hydrolase 2, N-(5,5-dioxodibenzothiophen-2-yl)-4,4-difluoro-piperidine-1-carboxamide | | Authors: | Dunge, A, Uwangue, O, Phan, C, Bjelcic, M, Gunnarsson, J, Wehlander, G, Kack, H, Branden, G. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring serial crystallography for drug discovery.
Iucrj, 2024
|
|
5Y59
 
 | | Crystal structure of Ku80 and Sir4 | | Descriptor: | ATP-dependent DNA helicase II subunit 2, SULFATE ION, Sir4p | | Authors: | Chen, H, Xue, J, Wu, J, Lei, M. | | Deposit date: | 2017-08-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural Insights into Yeast Telomerase Recruitment to Telomeres
Cell, 172, 2018
|
|
7WKP
 
 | | ICP1 Csy4 | | Descriptor: | 3' stem loop crRNA, Csy4 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
